Genomic clustering and homology between HET-S and the NWD2 STAND protein in various fungal genomes
- PMID: 22493719
- PMCID: PMC3321046
- DOI: 10.1371/journal.pone.0034854
Genomic clustering and homology between HET-S and the NWD2 STAND protein in various fungal genomes
Abstract
Background: Prions are infectious proteins propagating as self-perpetuating amyloid polymers. The [Het-s] prion of Podospora anserina is involved in a cell death process associated with non-self recognition. The prion forming domain (PFD) of HET-s adopts a β-solenoid amyloid structure characterized by the two fold repetition of an elementary triangular motif. [Het-s] induces cell death when interacting with HET-S, an allelic variant of HET-s. When templated by [Het-s], HET-S undergoes a trans-conformation, relocates to the cell membrane and induces toxicity.
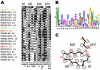
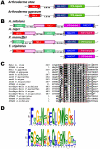
Methodology/principal findings: Here, comparing HET-s homologs from different species, we devise a consensus for the HET-s elementary triangular motif. We use this motif to screen genomic databases and find a match to the N-terminus of NWD2, a STAND protein, encoded by the gene immediately adjacent to het-S. STAND proteins are signal transducing ATPases which undergo ligand-induced oligomerisation. Homology modelling predicts that the NWD2 N-terminal region adopts a HET-s-like fold. We propose that upon NWD2 oligomerisation, these N-terminal extensions adopt the β-solenoid fold and template HET-S to adopt the amyloid fold and trigger toxicity. We extend this model to a putative prion, the σ infectious element in Nectria haematococca, because the s locus controlling propagation of σ also encodes a STAND protein and displays analogous features. Comparative genomic analyses indicate evolutionary conservation of these STAND/prion-like gene pairs, identify a number of novel prion candidates and define, in addition to the HET-s PFD motif, two distinct, novel putative PFD-like motifs.
Conclusions/significance: We suggest the existence, in the fungal kingdom, of a widespread and evolutionarily conserved mode of signal transduction based on the transmission of an amyloid-fold from a NOD-like STAND receptor protein to an effector protein.
Conflict of interest statement
Figures

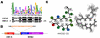






Similar articles
-
Signal transduction by a fungal NOD-like receptor based on propagation of a prion amyloid fold.PLoS Biol. 2015 Feb 11;13(2):e1002059. doi: 10.1371/journal.pbio.1002059. eCollection 2015 Feb. PLoS Biol. 2015. PMID: 25671553 Free PMC article.
-
Partial Prion Cross-Seeding between Fungal and Mammalian Amyloid Signaling Motifs.mBio. 2021 Feb 9;12(1):e02782-20. doi: 10.1128/mBio.02782-20. mBio. 2021. PMID: 33563842 Free PMC article.
-
The [Het-s] prion of Podospora anserina and its role in heterokaryon incompatibility.Semin Cell Dev Biol. 2011 Jul;22(5):460-8. doi: 10.1016/j.semcdb.2011.02.019. Epub 2011 Feb 18. Semin Cell Dev Biol. 2011. PMID: 21334447 Review.
-
Identification of a novel cell death-inducing domain reveals that fungal amyloid-controlled programmed cell death is related to necroptosis.Proc Natl Acad Sci U S A. 2016 Mar 8;113(10):2720-5. doi: 10.1073/pnas.1522361113. Epub 2016 Feb 22. Proc Natl Acad Sci U S A. 2016. PMID: 26903619 Free PMC article.
-
The HET-S/s Prion Motif in the Control of Programmed Cell Death.Cold Spring Harb Perspect Biol. 2016 Sep 1;8(9):a023515. doi: 10.1101/cshperspect.a023515. Cold Spring Harb Perspect Biol. 2016. PMID: 27352624 Free PMC article. Review.
Cited by
-
Signal transduction by a fungal NOD-like receptor based on propagation of a prion amyloid fold.PLoS Biol. 2015 Feb 11;13(2):e1002059. doi: 10.1371/journal.pbio.1002059. eCollection 2015 Feb. PLoS Biol. 2015. PMID: 25671553 Free PMC article.
-
Overlapping Podospora anserina Transcriptional Responses to Bacterial and Fungal Non Self Indicate a Multilayered Innate Immune Response.Front Microbiol. 2016 Apr 19;7:471. doi: 10.3389/fmicb.2016.00471. eCollection 2016. Front Microbiol. 2016. PMID: 27148175 Free PMC article.
-
Quantifying Nucleation In Vivo Reveals the Physical Basis of Prion-like Phase Behavior.Mol Cell. 2018 Jul 5;71(1):155-168.e7. doi: 10.1016/j.molcel.2018.06.016. Mol Cell. 2018. PMID: 29979963 Free PMC article.
-
Prion-like polymerization as a signaling mechanism.Trends Immunol. 2014 Dec;35(12):622-630. doi: 10.1016/j.it.2014.10.003. Epub 2014 Nov 12. Trends Immunol. 2014. PMID: 25457352 Free PMC article. Review.
-
Regulated Forms of Cell Death in Fungi.Front Microbiol. 2017 Sep 21;8:1837. doi: 10.3389/fmicb.2017.01837. eCollection 2017. Front Microbiol. 2017. PMID: 28983298 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

