Tudor staphylococcal nuclease (Tudor-SN) participates in small ribonucleoprotein (snRNP) assembly via interacting with symmetrically dimethylated Sm proteins
- PMID: 22493508
- PMCID: PMC3365748
- DOI: 10.1074/jbc.M111.311852
Tudor staphylococcal nuclease (Tudor-SN) participates in small ribonucleoprotein (snRNP) assembly via interacting with symmetrically dimethylated Sm proteins
Abstract
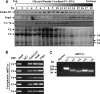
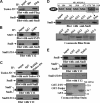
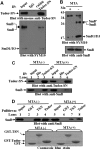
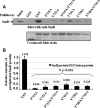
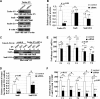
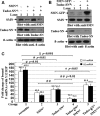
Human Tudor staphylococcal nuclease (Tudor-SN) is composed of four tandem repeats of staphylococcal nuclease (SN)-like domains, followed by a tudor and SN-like domain (TSN) consisting of a central tudor flanked by two partial SN-like sequences. The crystal structure of the tudor domain displays a conserved aromatic cage, which is predicted to hook methyl groups. Here, we demonstrated that the TSN domain of Tudor-SN binds to symmetrically dimethylarginine (sDMA)-modified SmB/B' and SmD1/D3 core proteins of the spliceosome. We demonstrated that this interaction ability is reduced by the methyltransferase inhibitor 5-deoxy-5-(methylthio)adenosine. Mutagenesis experiments indicated that the conserved amino acids (Phe-715, Tyr-721, Tyr-738, and Tyr-741) in the methyl-binding cage of the TSN domain are required for Tudor-SN-SmB interaction. Furthermore, depletion of Tudor-SN affects the association of Sm protein with snRNAs and, as a result, inhibits the assembly of uridine-rich small ribonucleoprotein mediated by the Sm core complex in vivo. Our results reveal the molecular basis for the involvement of Tudor-SN in regulating small nuclear ribonucleoprotein biogenesis, which provides novel insight related to the biological activity of Tudor-SN.
Figures
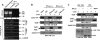


Similar articles
-
Transcriptional co-activator protein p100 interacts with snRNP proteins and facilitates the assembly of the spliceosome.Nucleic Acids Res. 2007;35(13):4485-94. doi: 10.1093/nar/gkm470. Epub 2007 Jun 18. Nucleic Acids Res. 2007. PMID: 17576664 Free PMC article.
-
Structure and ligand binding of the extended Tudor domain of D. melanogaster Tudor-SN.J Mol Biol. 2009 Apr 10;387(4):921-34. doi: 10.1016/j.jmb.2009.02.018. Epub 2009 Feb 14. J Mol Biol. 2009. PMID: 19232356
-
Plasmodium falciparum Tudor Staphylococcal Nuclease interacting proteins suggest its role in nuclear as well as splicing processes.Gene. 2010 Nov 15;468(1-2):48-57. doi: 10.1016/j.gene.2010.08.004. Epub 2010 Aug 14. Gene. 2010. PMID: 20713134
-
Tudor staphylococcal nuclease: biochemistry and functions.Cell Death Differ. 2016 Nov 1;23(11):1739-1748. doi: 10.1038/cdd.2016.93. Epub 2016 Sep 9. Cell Death Differ. 2016. PMID: 27612014 Free PMC article. Review.
-
The SMN complex: an assembly machine for RNPs.Cold Spring Harb Symp Quant Biol. 2006;71:313-20. doi: 10.1101/sqb.2006.71.001. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381311 Review.
Cited by
-
SND1 acts as an anti-apoptotic factor via regulating the expression of lncRNA UCA1 in hepatocellular carcinoma.RNA Biol. 2018;15(10):1364-1375. doi: 10.1080/15476286.2018.1534525. Epub 2018 Oct 25. RNA Biol. 2018. PMID: 30321081 Free PMC article.
-
Tudor staphylococcal nuclease links formation of stress granules and processing bodies with mRNA catabolism in Arabidopsis.Plant Cell. 2015 Mar;27(3):926-43. doi: 10.1105/tpc.114.134494. Epub 2015 Mar 3. Plant Cell. 2015. PMID: 25736060 Free PMC article.
-
Recruitment of the NineTeen Complex to the activated spliceosome requires AtPRMT5.Proc Natl Acad Sci U S A. 2016 May 10;113(19):5447-52. doi: 10.1073/pnas.1522458113. Epub 2016 Apr 25. Proc Natl Acad Sci U S A. 2016. PMID: 27114555 Free PMC article.
-
Organization of Plasmodium falciparum spliceosomal core complex and role of arginine methylation in its assembly.Malar J. 2013 Sep 18;12:333. doi: 10.1186/1475-2875-12-333. Malar J. 2013. PMID: 24047207 Free PMC article.
-
Contribution of alternative splicing to breast cancer metastasis.J Cancer Metastasis Treat. 2019;5:21. doi: 10.20517/2394-4722.2018.96. Epub 2019 Mar 22. J Cancer Metastasis Treat. 2019. PMID: 31737791 Free PMC article.
References
-
- Broadhurst M. K., Lee R. S., Hawkins S., Wheeler T. T. (2005) The p100 EBNA-2 co-activator. A highly conserved protein found in a range of exocrine and endocrine cells and tissues in cattle. Biochim. Biophys. Acta 1681, 126–133 - PubMed
-
- Rodríguez L., Ochoa B., Martínez M. J. (2007) NF-Y and Sp1 are involved in transcriptional regulation of rat SND p102 gene. Biochem. Biophys. Res. Commun. 356, 226–232 - PubMed
-
- Zhao C. T., Shi K. H., Su Y., Liang L. Y., Yan Y., Postlethwait J., Meng A.M. (2003) Two variants of zebrafish p100 are expressed during embryogenesis and regulated by Nodal signaling. FEBS Lett. 543, 190–195 - PubMed
-
- Sami-Subbu R., Choi S. B., Wu Y., Wang C., Okita T. W. (2001) Identification of a cytoskeleton-associated 120-kDa RNA-binding protein in developing rice seeds. Plant Mol. Biol. 46, 79–88 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

