Interaction of chandipura virus N and P proteins: identification of two mutually exclusive domains of N involved in interaction with P
- PMID: 22485180
- PMCID: PMC3317646
- DOI: 10.1371/journal.pone.0034623
Interaction of chandipura virus N and P proteins: identification of two mutually exclusive domains of N involved in interaction with P
Abstract
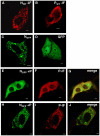
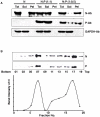
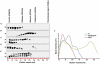
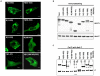
The nucleocapsid protein (N) and the phosphoprotein (P) of nonsegmented negative-strand (NNS) RNA viruses interact with each other to accomplish two crucial events necessary for the viral replication cycle. First, the P protein binds to the aggregation prone nascent N molecules maintaining them in a soluble monomeric (N(0)) form (N(0)-P complex). It is this form that is competent for specific encapsidation of the viral genome. Second, the P protein binds to oligomeric N in the nucleoprotein complex (N-RNA-P complex), and thereby facilitates the recruitment of the viral polymerase (L) onto its template. All previous attempts to study these complexes relied on co-expression of the two proteins in diverse systems. In this study, we have characterised these different modes of N-P interaction in detail and for the first time have been able to reconstitute these complexes individually in vitro in the chandipura virus (CHPV), a human pathogenic NNS RNA virus. Using a battery of truncated mutants of the N protein, we have been able to identify two mutually exclusive domains of N involved in differential interaction with the P protein. An unique N-terminal binding site, comprising of amino acids (aa) 1-180 form the N(0)-P interacting region, whereas, C-terminal residues spanning aa 320-390 is instrumental in N-RNA-P interactions. Significantly, the ex-vivo data also supports these observations. Based on these results, we suggest that the P protein acts as N-specific chaperone and thereby partially masking the N-N self-association region, which leads to the specific recognition of viral genome RNA by N(0).
Conflict of interest statement
Figures


Similar articles
-
Interaction of vesicular stomatitis virus P and N proteins: identification of two overlapping domains at the N terminus of P that are involved in N0-P complex formation and encapsidation of viral genome RNA.J Virol. 2007 Dec;81(24):13478-85. doi: 10.1128/JVI.01244-07. Epub 2007 Oct 3. J Virol. 2007. PMID: 17913815 Free PMC article.
-
Oligomerization of the Vesicular Stomatitis Virus Phosphoprotein Is Dispensable for mRNA Synthesis but Facilitates RNA Replication.J Virol. 2020 Jun 16;94(13):e00115-20. doi: 10.1128/JVI.00115-20. Print 2020 Jun 16. J Virol. 2020. PMID: 32321813 Free PMC article.
-
Characterization of the chandipura virus leader RNA-phosphoprotein interaction using single tryptophan mutants and its detection in viral infected cells.Biochimie. 2013 Feb;95(2):180-94. doi: 10.1016/j.biochi.2012.09.009. Epub 2012 Oct 9. Biochimie. 2013. PMID: 23063516
-
Reviewing Chandipura: a vesiculovirus in human epidemics.Biosci Rep. 2007 Oct;27(4-5):275-98. doi: 10.1007/s10540-007-9054-z. Biosci Rep. 2007. PMID: 17610154 Free PMC article. Review.
-
Interactions between the Nucleoprotein and the Phosphoprotein of Pneumoviruses: Structural Insight for Rational Design of Antivirals.Viruses. 2021 Dec 6;13(12):2449. doi: 10.3390/v13122449. Viruses. 2021. PMID: 34960719 Free PMC article. Review.
Cited by
-
Host-rabies virus protein-protein interactions as druggable antiviral targets.Proc Natl Acad Sci U S A. 2013 Mar 5;110(10):E861-8. doi: 10.1073/pnas.1210198110. Epub 2013 Feb 12. Proc Natl Acad Sci U S A. 2013. PMID: 23404707 Free PMC article.
-
Phosphorylation at the homotypic interface regulates nucleoprotein oligomerization and assembly of the influenza virus replication machinery.PLoS Pathog. 2015 Apr 13;11(4):e1004826. doi: 10.1371/journal.ppat.1004826. eCollection 2015 Apr. PLoS Pathog. 2015. PMID: 25867750 Free PMC article.
-
Elucidating the interacting domains of chandipura virus nucleocapsid protein.Adv Virol. 2013;2013:594319. doi: 10.1155/2013/594319. Epub 2013 Oct 28. Adv Virol. 2013. PMID: 24288532 Free PMC article.
-
Newly identified phosphorylation site in the vesicular stomatitis virus P protein is required for viral RNA synthesis.J Virol. 2014 Feb;88(3):1461-72. doi: 10.1128/JVI.02384-13. Epub 2013 Nov 20. J Virol. 2014. PMID: 24257610 Free PMC article.
-
Analysis of the dark proteome of Chandipura virus reveals maximum propensity for intrinsic disorder in phosphoprotein.Sci Rep. 2021 Jun 24;11(1):13253. doi: 10.1038/s41598-021-92581-6. Sci Rep. 2021. PMID: 34168211 Free PMC article.
References
-
- Bhatt PN, Rodrigues FM. Chandipura: a new Arbovirus isolated in India from patients with febrile illness. Indian J Med Res. 1967;55:1295–1305. - PubMed
-
- Chadha MS, Arankalle VA, Jadi RS, Joshi MV, Thakare JP, et al. An outbreak of Chandipura virus encephalitis in the eastern districts of Gujarat state, India. Am J Trop Med Hyg. 2005;73:566–570. - PubMed
-
- Gurav YK, Tandale BV, Jadi RS, Gunjikar RS, Tikute SS, et al. Chandipura virus encephalitis outbreak among children in Nagpur division, Maharashtra, 2007. Indian J Med Res. 2010;132:395–399. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

