Iron sulfur cluster proteins and microbial regulation: implications for understanding tuberculosis
- PMID: 22483328
- PMCID: PMC3962503
- DOI: 10.1016/j.cbpa.2012.03.004
Iron sulfur cluster proteins and microbial regulation: implications for understanding tuberculosis
Abstract
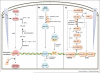
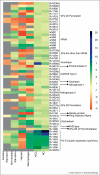
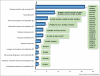
All pathogenic and nonpathogenic microbes are continuously exposed to environmental or endogenous reactive oxygen and nitrogen species, which can critically effect survival and disease. Iron-sulfur [Fe-S] cluster containing prosthetic groups provide the microbial cell with a unique capacity to sense and transcriptionally respond to diatomic gases (e.g. NO and O2) and redox-cycling agents. Recent advances in our understanding of the mechanisms for how the FNR and SoxR [Fe-S] cluster proteins respond to NO and O2 have provided new insights into the biochemical mechanism of action of the Mycobacterium tuberculosis (Mtb) family of WhiB [Fe-S] cluster proteins. These insights have provided the basis for establishing a unifying paradigm for the Mtb WhiB family of proteins. Mtb is the etiological agent for tuberculosis (TB), a disease that affects nearly one-third of the world's population.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures
Similar articles
-
Intracellular peroxynitrite perturbs redox balance, bioenergetics, and Fe-S cluster homeostasis in Mycobacterium tuberculosis.Redox Biol. 2024 Sep;75:103285. doi: 10.1016/j.redox.2024.103285. Epub 2024 Jul 31. Redox Biol. 2024. PMID: 39128229 Free PMC article. Review.
-
Mycobacterium tuberculosis WhiB3 responds to O2 and nitric oxide via its [4Fe-4S] cluster and is essential for nutrient starvation survival.Proc Natl Acad Sci U S A. 2007 Jul 10;104(28):11562-7. doi: 10.1073/pnas.0700490104. Epub 2007 Jul 3. Proc Natl Acad Sci U S A. 2007. PMID: 17609386 Free PMC article.
-
Mycobacterium tuberculosis WhiB3: a novel iron-sulfur cluster protein that regulates redox homeostasis and virulence.Antioxid Redox Signal. 2012 Apr 1;16(7):687-97. doi: 10.1089/ars.2011.4341. Antioxid Redox Signal. 2012. PMID: 22010944 Free PMC article. Review.
-
Structural insights into the functional divergence of WhiB-like proteins in Mycobacterium tuberculosis.Mol Cell. 2021 Jul 15;81(14):2887-2900.e5. doi: 10.1016/j.molcel.2021.06.002. Epub 2021 Jun 24. Mol Cell. 2021. PMID: 34171298 Free PMC article.
-
Studies on structural and functional divergence among seven WhiB proteins of Mycobacterium tuberculosis H37Rv.FEBS J. 2009 Jan;276(1):76-93. doi: 10.1111/j.1742-4658.2008.06755.x. FEBS J. 2009. PMID: 19016840
Cited by
-
Causal Relationship between Mitochondrial-Associated Proteins and Sepsis in ICU Patients: A Mendelian Randomization Study.ACS Omega. 2024 Feb 6;9(7):8457-8463. doi: 10.1021/acsomega.3c09676. eCollection 2024 Feb 20. ACS Omega. 2024. PMID: 38405532 Free PMC article.
-
The Multifaceted Bacterial Cysteine Desulfurases: From Metabolism to Pathogenesis.Antioxidants (Basel). 2021 Jun 23;10(7):997. doi: 10.3390/antiox10070997. Antioxidants (Basel). 2021. PMID: 34201508 Free PMC article. Review.
-
Intracellular peroxynitrite perturbs redox balance, bioenergetics, and Fe-S cluster homeostasis in Mycobacterium tuberculosis.Redox Biol. 2024 Sep;75:103285. doi: 10.1016/j.redox.2024.103285. Epub 2024 Jul 31. Redox Biol. 2024. PMID: 39128229 Free PMC article. Review.
-
Host-pathogen redox dynamics modulate Mycobacterium tuberculosis pathogenesis.Pathog Dis. 2018 Jul 1;76(5):fty036. doi: 10.1093/femspd/fty036. Pathog Dis. 2018. PMID: 29873719 Free PMC article.
-
WhiB6 regulation of ESX-1 gene expression is controlled by a negative feedback loop in Mycobacterium marinum.Proc Natl Acad Sci U S A. 2017 Dec 12;114(50):E10772-E10781. doi: 10.1073/pnas.1710167114. Epub 2017 Nov 27. Proc Natl Acad Sci U S A. 2017. PMID: 29180415 Free PMC article.
References
-
- Py B, Moreau PL, Barras F. Fe–S clusters, fragile sentinels of the cell. Curr Opin Microbiol. 2011;14:218–223. - PubMed
-
- Xu XM, Moller SG. Iron sulfur clusters: biogenesis, molecular mechanisms, and their functional significance. Antioxid Redox Signal. 2011;15:271–307. - PubMed
-
- Liu Y, Sieprawska-Lupa M, Whitman WB, White RH. Cysteine is not the sulfur source for iron-sulfur cluster and methionine biosynthesis in the methanogenic archaeon Methanococcus maripaludis. J Biol Chem. 2010;285:31923–31929. [This is the first demonstration that sulfide can be used as a source of sulfur during Fe–S cluster biosynthesis. Usually organisms that stay in sulfide-rich habitats, mostly those belonging to archaea, use sulfide rather than cysteine to make Fe–S clusters.] - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

