Differential transcription factor use by the KIR2DL4 promoter under constitutive and IL-2/15-treated conditions
- PMID: 22467658
- PMCID: PMC3331908
- DOI: 10.4049/jimmunol.1103352
Differential transcription factor use by the KIR2DL4 promoter under constitutive and IL-2/15-treated conditions
Abstract
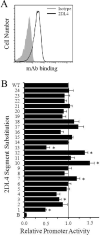
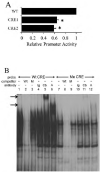
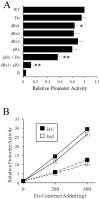
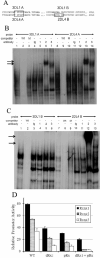
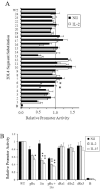
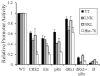
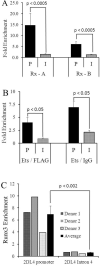
KIR2DL4 is unique among human KIR genes in expression, cellular localization, structure, and function, yet the transcription factors required for its expression have not been identified. Using mutagenesis, EMSA, and cotransfection assays, we identified two redundant Runx binding sites in the 2DL4 promoter as essential for constitutive 2DL4 transcription, with contributions by a cyclic AMP response element (CRE) and initiator elements. IL-2- and IL-15-stimulated human NK cell lines increased 2DL4 promoter activity, which required functional Runx, CRE, and Ets sites. Chromatin immunoprecipitation experiments show that Runx3 and Ets1 bind the 2DL4 promoter in situ. 2DL4 promoter activity had similar transcription factor requirements in T cells. Runx, CRE, and Ets binding motifs are present in 2DL4 promoters from across primate species, but other postulated transcription factor binding sites are not preserved. Differences between 2DL4 and clonally restricted KIR promoters suggest a model that explains the unique 2DL4 expression pattern in human NK cells.
Figures
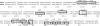

Similar articles
-
Human CD94 gene expression: dual promoters differing in responsiveness to IL-2 or IL-15.J Immunol. 2003 Nov 15;171(10):5277-86. doi: 10.4049/jimmunol.171.10.5277. J Immunol. 2003. PMID: 14607929
-
KIR2DL4 is an IL-2-regulated NK cell receptor that exhibits limited expression in humans but triggers strong IFN-gamma production.J Immunol. 2003 Oct 1;171(7):3415-25. doi: 10.4049/jimmunol.171.7.3415. J Immunol. 2003. PMID: 14500636
-
The transcription factor Ets1 is important for CD4 repression and Runx3 up-regulation during CD8 T cell differentiation in the thymus.J Exp Med. 2009 Nov 23;206(12):2685-99. doi: 10.1084/jem.20092024. Epub 2009 Nov 16. J Exp Med. 2009. PMID: 19917777 Free PMC article.
-
Roles of HLA-G/KIR2DL4 in Breast Cancer Immune Microenvironment.Front Immunol. 2022 Feb 3;13:791975. doi: 10.3389/fimmu.2022.791975. eCollection 2022. Front Immunol. 2022. PMID: 35185887 Free PMC article. Review.
-
Killer Immunoglobulin-Like Receptor 2DL4 (CD158d) Regulates Human Mast Cells both Positively and Negatively: Possible Roles in Pregnancy and Cancer Metastasis.Int J Mol Sci. 2020 Jan 31;21(3):954. doi: 10.3390/ijms21030954. Int J Mol Sci. 2020. PMID: 32023940 Free PMC article. Review.
Cited by
-
Transcription factor Runx3 regulates interleukin-15-dependent natural killer cell activation.Mol Cell Biol. 2014 Mar;34(6):1158-69. doi: 10.1128/MCB.01202-13. Epub 2014 Jan 13. Mol Cell Biol. 2014. PMID: 24421391 Free PMC article.
-
Killer-cell immunoglobulin-like receptor genes and ligands and their role in hematologic malignancies.Cancer Immunol Immunother. 2016 Apr;65(4):427-40. doi: 10.1007/s00262-016-1806-9. Epub 2016 Feb 13. Cancer Immunol Immunother. 2016. PMID: 26874942 Free PMC article. Review.
-
Runx3-mediated transcriptional program in cytotoxic lymphocytes.PLoS One. 2013 Nov 13;8(11):e80467. doi: 10.1371/journal.pone.0080467. eCollection 2013. PLoS One. 2013. PMID: 24236182 Free PMC article.
-
Transcriptional and post-transcriptional regulation of NK cell development and function.Clin Immunol. 2017 Apr;177:60-69. doi: 10.1016/j.clim.2016.03.003. Epub 2016 Mar 3. Clin Immunol. 2017. PMID: 26948928 Free PMC article. Review.
-
IL-2/IL-15 activate the human clonally restricted KIR3DL1 reverse promoter.Genes Immun. 2013 Mar;14(2):107-14. doi: 10.1038/gene.2012.62. Epub 2013 Jan 17. Genes Immun. 2013. PMID: 23328843 Free PMC article.
References
-
- Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. - PubMed
-
- Rajalingam R, Parham P, Abi-Rached L. Domain shuffling has been the main mechanism forming new hominoid killer cell Ig-like receptors. J Immunol. 2004;172:356–369. - PubMed
-
- Santourlidis S, Graffmann N, Christ J, Uhrberg M. Lineage-specific transition of histone signatures in the killer cell Ig-like receptor locus from hematopoietic progenitor to NK cells. J Immunol. 2008;180:418–425. - PubMed
-
- Santourlidis S, Trompeter HI, Weinhold S, Eisermann B, Meyer KL, Wernet P, Uhrberg M. Crucial role of DNA methylation in determination of clonally distributed killer cell Ig-like receptor expression patterns in NK cells. J Immunol. 2002;169:4253–4261. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

