Transcriptional profiling of stroma from inflamed and resting lymph nodes defines immunological hallmarks
- PMID: 22466668
- PMCID: PMC3366863
- DOI: 10.1038/ni.2262
Transcriptional profiling of stroma from inflamed and resting lymph nodes defines immunological hallmarks
Abstract
Lymph node stromal cells (LNSCs) closely regulate immunity and self-tolerance, yet key aspects of their biology remain poorly elucidated. Here, comparative transcriptomic analyses of mouse LNSC subsets demonstrated the expression of important immune mediators, growth factors and previously unknown structural components. Pairwise analyses of ligands and cognate receptors across hematopoietic and stromal subsets suggested a complex web of crosstalk. Fibroblastic reticular cells (FRCs) showed enrichment for higher expression of genes relevant to cytokine signaling, relative to their expression in skin and thymic fibroblasts. LNSCs from inflamed lymph nodes upregulated expression of genes encoding chemokines and molecules involved in the acute-phase response and the antigen-processing and antigen-presentation machinery. Poorly studied podoplanin (gp38)-negative CD31(-) LNSCs showed similarities to FRCs but lacked expression of interleukin 7 (IL-7) and were identified as myofibroblastic pericytes that expressed integrin α(7). Together our data comprehensively describe the transcriptional characteristics of LNSC subsets.
Figures

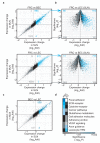



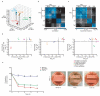
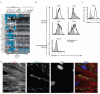

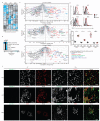
Similar articles
-
Lymph node fibroblastic reticular cells directly present peripheral tissue antigen under steady-state and inflammatory conditions.J Exp Med. 2010 Apr 12;207(4):689-97. doi: 10.1084/jem.20092642. Epub 2010 Mar 22. J Exp Med. 2010. PMID: 20308362 Free PMC article.
-
SIRPα+ dendritic cells promote the development of fibroblastic reticular cells in murine peripheral lymph nodes.Eur J Immunol. 2019 Sep;49(9):1364-1371. doi: 10.1002/eji.201948103. Epub 2019 Jun 3. Eur J Immunol. 2019. PMID: 31099900
-
Lymph node stromal cells negatively regulate antigen-specific CD4+ T cell responses.J Immunol. 2014 Aug 15;193(4):1636-44. doi: 10.4049/jimmunol.1302946. Epub 2014 Jul 14. J Immunol. 2014. PMID: 25024385
-
Lymph node stroma broaden the peripheral tolerance paradigm.Trends Immunol. 2011 Jan;32(1):12-8. doi: 10.1016/j.it.2010.11.002. Epub 2010 Dec 10. Trends Immunol. 2011. PMID: 21147035 Free PMC article. Review.
-
Peripheral tolerance induction by lymph node stroma.Adv Exp Med Biol. 2009;633:113-27. doi: 10.1007/978-0-387-79311-5_10. Adv Exp Med Biol. 2009. PMID: 19209685 Review.
Cited by
-
BatchFLEX: feature-level equalization of X-batch.Bioinformatics. 2024 Oct 1;40(10):btae587. doi: 10.1093/bioinformatics/btae587. Bioinformatics. 2024. PMID: 39360977 Free PMC article.
-
IL4RA on lymphatic endothelial cells promotes T cell egress during sclerodermatous graft versus host disease.JCI Insight. 2016 Aug 4;1(12):e88057. doi: 10.1172/jci.insight.88057. JCI Insight. 2016. PMID: 27547823 Free PMC article.
-
Dissection of DLBCL microenvironment provides a gene expression-based predictor of survival applicable to formalin-fixed paraffin-embedded tissue.Ann Oncol. 2018 Dec 1;29(12):2363-2370. doi: 10.1093/annonc/mdy450. Ann Oncol. 2018. PMID: 30307529 Free PMC article.
-
Distinctive expression of T cell guiding molecules in human autoimmune lymph node stromal cells upon TLR3 triggering.Sci Rep. 2018 Jan 29;8(1):1736. doi: 10.1038/s41598-018-19951-5. Sci Rep. 2018. PMID: 29379035 Free PMC article.
-
A Single-Cell Transcriptional Roadmap of the Mouse and Human Lymph Node Lymphatic Vasculature.Front Cardiovasc Med. 2020 Apr 30;7:52. doi: 10.3389/fcvm.2020.00052. eCollection 2020. Front Cardiovasc Med. 2020. PMID: 32426372 Free PMC article.
References
-
- Heng TS, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi:10.1038/ni1008-1091. - PubMed
-
- Link A, et al. Fibroblastic reticular cells in lymph nodes regulate the homeostasis of naive T cells. Nat Immunol. 2007;8:1255–1265. doi:10.1038/ni1513. - PubMed
-
- Lee JW, et al. Peripheral antigen display by lymph node stroma promotes T cell tolerance to intestinal self. Nat Immunol. 2007;8:181–190. doi:10.1038/ni1427. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- R01 DK074500/DK/NIDDK NIH HHS/United States
- GM38903/GM/NIGMS NIH HHS/United States
- R01 CA154947/CA/NCI NIH HHS/United States
- R01 DE019917/DE/NIDCR NIH HHS/United States
- R01 AI063428/AI/NIAID NIH HHS/United States
- R01 AI063428-06/AI/NIAID NIH HHS/United States
- P01 AI045757/AI/NIAID NIH HHS/United States
- R01 AI072117/AI/NIAID NIH HHS/United States
- R24 AI072073/AI/NIAID NIH HHS/United States
- R37 AI067545/AI/NIAID NIH HHS/United States
- R01 GM038903-25/GM/NIGMS NIH HHS/United States
- R01 GM038903/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

