c-Jun N-terminal kinase-mediated Rad18 phosphorylation facilitates Polη recruitment to stalled replication forks
- PMID: 22456510
- PMCID: PMC3350557
- DOI: 10.1091/mbc.E11-10-0829
c-Jun N-terminal kinase-mediated Rad18 phosphorylation facilitates Polη recruitment to stalled replication forks
Abstract
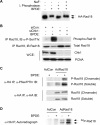
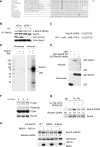
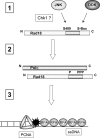
The E3 ubiquitin ligase Rad18 chaperones DNA polymerase η (Polη) to sites of UV-induced DNA damage and monoubiquitinates proliferating cell nuclear antigen (PCNA), facilitating engagement of Polη with stalled replication forks and promoting translesion synthesis (TLS). It is unclear how Rad18 activities are coordinated with other elements of the DNA damage response. We show here that Ser-409 residing in the Polη-binding motif of Rad18 is phosphorylated in a checkpoint kinase 1-dependent manner in genotoxin-treated cells. Recombinant Rad18 was phosphorylated specifically at S409 by c-Jun N-terminal kinase (JNK) in vitro. In UV-treated cells, Rad18 S409 phosphorylation was inhibited by a pharmacological JNK inhibitor. Conversely, ectopic expression of JNK and its upstream kinase mitogen-activated protein kinase kinase 4 led to DNA damage-independent Rad18 S409 phosphorylation. These results identify Rad18 as a novel JNK substrate. A Rad18 mutant harboring a Ser → Ala substitution at S409 was compromised for Polη association and did not redistribute Polη to nuclear foci or promote Polη-PCNA interaction efficiently relative to wild-type Rad18. Rad18 S409A also failed to fully complement the UV sensitivity of Rad18-depleted cells. Taken together, these results show that Rad18 phosphorylation by JNK represents a novel mechanism for promoting TLS and DNA damage tolerance.
Figures



Similar articles
-
A non-catalytic role of DNA polymerase η in recruiting Rad18 and promoting PCNA monoubiquitination at stalled replication forks.Nucleic Acids Res. 2013 Mar 1;41(5):3079-93. doi: 10.1093/nar/gkt016. Epub 2013 Jan 23. Nucleic Acids Res. 2013. PMID: 23345618 Free PMC article.
-
Phosphorylated Rad18 directs DNA polymerase η to sites of stalled replication.J Cell Biol. 2010 Nov 29;191(5):953-66. doi: 10.1083/jcb.201006043. Epub 2010 Nov 22. J Cell Biol. 2010. PMID: 21098111 Free PMC article.
-
Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination.EMBO J. 2004 Oct 1;23(19):3886-96. doi: 10.1038/sj.emboj.7600383. Epub 2004 Sep 9. EMBO J. 2004. PMID: 15359278 Free PMC article.
-
Integrating DNA replication with trans-lesion synthesis via Cdc7.Cell Cycle. 2010 Dec 15;9(24):4818-23. doi: 10.4161/cc.9.24.14241. Epub 2010 Dec 15. Cell Cycle. 2010. PMID: 21150323 Review.
-
Regulation of DNA damage tolerance in mammalian cells by post-translational modifications of PCNA.Mutat Res. 2017 Oct;803-805:82-88. doi: 10.1016/j.mrfmmm.2017.06.004. Epub 2017 Jun 21. Mutat Res. 2017. PMID: 28666590 Review.
Cited by
-
Control of DNA Damage Bypass by Ubiquitylation of PCNA.Genes (Basel). 2020 Jan 29;11(2):138. doi: 10.3390/genes11020138. Genes (Basel). 2020. PMID: 32013080 Free PMC article. Review.
-
Roles of trans-lesion synthesis (TLS) DNA polymerases in tumorigenesis and cancer therapy.NAR Cancer. 2023 Feb 6;5(1):zcad005. doi: 10.1093/narcan/zcad005. eCollection 2023 Mar. NAR Cancer. 2023. PMID: 36755961 Free PMC article.
-
The Global Phosphorylation Landscape of SARS-CoV-2 Infection.Cell. 2020 Aug 6;182(3):685-712.e19. doi: 10.1016/j.cell.2020.06.034. Epub 2020 Jun 28. Cell. 2020. PMID: 32645325 Free PMC article.
-
A non-catalytic role of DNA polymerase η in recruiting Rad18 and promoting PCNA monoubiquitination at stalled replication forks.Nucleic Acids Res. 2013 Mar 1;41(5):3079-93. doi: 10.1093/nar/gkt016. Epub 2013 Jan 23. Nucleic Acids Res. 2013. PMID: 23345618 Free PMC article.
-
Overexpression of oncogenic H-Ras in hTERT-immortalized and SV40-transformed human cells targets replicative and specialized DNA polymerases for depletion.PLoS One. 2021 May 7;16(5):e0251188. doi: 10.1371/journal.pone.0251188. eCollection 2021. PLoS One. 2021. PMID: 33961649 Free PMC article.
References
-
- Bi X, Slater DM, Ohmori H, Vaziri C. DNA polymerase kappa is specifically required for recovery from the benzo[a]pyrene-dihydrodiol epoxide (BPDE)-induced S-phase checkpoint. J Biol Chem. 2005;280:22343–22355. - PubMed
-
- Bienko M, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

