Cohesin, CTCF and lymphocyte antigen receptor locus rearrangement
- PMID: 22440186
- PMCID: PMC3352889
- DOI: 10.1016/j.it.2012.02.004
Cohesin, CTCF and lymphocyte antigen receptor locus rearrangement
Abstract
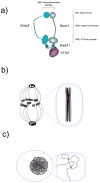
The somatic recombination of lymphocyte antigen receptor loci is integral to lymphocyte differentiation and adaptive immunity. Here we review the relation of this highly choreographed process with the zinc finger protein CTCF and with cohesin, a protein complex best known for its essential functions in post-replicative DNA repair and chromosome segregation during the cell cycle. At lymphocyte antigen receptor loci, CTCF and cohesin shape long-range interactions and contribute to V(D)J recombination by facilitating lineage- and developmental-stage-specific transcription and accessibility.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Conflict of interest statement
The authors have no financial conflict of interest.
Figures




Similar articles
-
Variable Extent of Lineage-Specificity and Developmental Stage-Specificity of Cohesin and CCCTC-Binding Factor Binding Within the Immunoglobulin and T Cell Receptor Loci.Front Immunol. 2018 Mar 8;9:425. doi: 10.3389/fimmu.2018.00425. eCollection 2018. Front Immunol. 2018. PMID: 29593713 Free PMC article.
-
Cutting edge: developmental stage-specific recruitment of cohesin to CTCF sites throughout immunoglobulin loci during B lymphocyte development.J Immunol. 2009 Jan 1;182(1):44-8. doi: 10.4049/jimmunol.182.1.44. J Immunol. 2009. PMID: 19109133 Free PMC article.
-
CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells.Proc Natl Acad Sci U S A. 2011 Jun 7;108(23):9566-71. doi: 10.1073/pnas.1019391108. Epub 2011 May 23. Proc Natl Acad Sci U S A. 2011. PMID: 21606361 Free PMC article.
-
The role of CTCF in regulating V(D)J recombination.Curr Opin Immunol. 2012 Apr;24(2):153-9. doi: 10.1016/j.coi.2012.01.003. Epub 2012 Mar 15. Curr Opin Immunol. 2012. PMID: 22424610 Free PMC article. Review.
-
DNA-binding factor CTCF and long-range gene interactions in V(D)J recombination and oncogene activation.Blood. 2012 Jun 28;119(26):6209-18. doi: 10.1182/blood-2012-03-402586. Epub 2012 Apr 26. Blood. 2012. PMID: 22538856 Review.
Cited by
-
Sister chromatid cohesion.Cold Spring Harb Perspect Biol. 2012 Nov 1;4(11):a011130. doi: 10.1101/cshperspect.a011130. Cold Spring Harb Perspect Biol. 2012. PMID: 23043155 Free PMC article. Review.
-
Complete cis Exclusion upon Duplication of the Eμ Enhancer at the Immunoglobulin Heavy Chain Locus.Mol Cell Biol. 2015 Jul;35(13):2231-41. doi: 10.1128/MCB.00294-15. Mol Cell Biol. 2015. PMID: 25896912 Free PMC article.
-
Cohesin and polycomb proteins functionally interact to control transcription at silenced and active genes.PLoS Genet. 2013 Jun;9(6):e1003560. doi: 10.1371/journal.pgen.1003560. Epub 2013 Jun 20. PLoS Genet. 2013. PMID: 23818863 Free PMC article.
-
Epigenetics and the adaptive immune response.Mol Aspects Med. 2013 Jul-Aug;34(4):813-25. doi: 10.1016/j.mam.2012.06.008. Epub 2012 Jul 10. Mol Aspects Med. 2013. PMID: 22789989 Free PMC article. Review.
-
Regulatory Network of PD1 Signaling Is Associated with Prognosis in Glioblastoma Multiforme.Cancer Res. 2021 Nov 1;81(21):5401-5412. doi: 10.1158/0008-5472.CAN-21-0730. Epub 2021 Sep 7. Cancer Res. 2021. PMID: 34493595 Free PMC article.
References
-
- Schatz DG, Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nat Rev Immunol. 2011;11:251–263. - PubMed
-
- Skok JA, Brown KE, Azuara V, Caparros ML, Baxter J, Takacs K, Dillon N, Gray D, Perry RP, Merkenschlager M, Fisher AG. Nonequivalent nuclear location of immunoglobulin alleles in B lymphocytes. Nat Immunol. 2001;2:848–854. - PubMed
-
- Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

