Keratinocyte growth factor induces gene expression signature associated with suppression of malignant phenotype of cutaneous squamous carcinoma cells
- PMID: 22427941
- PMCID: PMC3299721
- DOI: 10.1371/journal.pone.0033041
Keratinocyte growth factor induces gene expression signature associated with suppression of malignant phenotype of cutaneous squamous carcinoma cells
Abstract
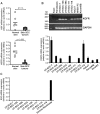
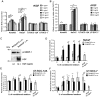
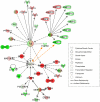
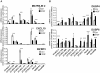
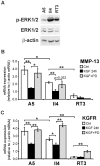
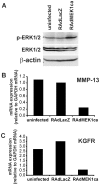
Keratinocyte growth factor (KGF, fibroblast growth factor-7) is a fibroblast-derived mitogen, which stimulates proliferation of epithelial cells. The expression of KGF by dermal fibroblasts is induced following injury and it promotes wound repair. However, the role of KGF in cutaneous carcinogenesis and cancer progression is not known. We have examined the role of KGF in progression of squamous cell carcinoma (SCC) of the skin. The expression of KGF receptor (KGFR) mRNA was lower in cutaneous SCCs (n = 6) than in normal skin samples (n = 6). Expression of KGFR mRNA was detected in 6 out of 8 cutaneous SCC cell lines and the levels were downregulated by 24-h treatment with KGF. KGF did not stimulate SCC cell proliferation, but it reduced invasion of SCC cells through collagen. Gene expression profiling of three cutaneous SCC cell lines treated with KGF for 24 h revealed a specific gene expression signature characterized by upregulation of a set of genes specifically downregulated in SCC cells compared to normal epidermal keratinocytes, including genes with tumor suppressing properties (SPRY4, DUSP4, DUSP6, LRIG1, PHLDA1). KGF also induced downregulation of a set of genes specifically upregulated in SCC cells compared to normal keratinocytes, including genes associated with tumor progression (MMP13, MATN2, CXCL10, and IGFBP3). Downregulation of MMP-13 and KGFR expression in SCC cells and HaCaT cells was mediated via ERK1/2. Activation of ERK1/2 in HaCaT cells and tumorigenic Ha-ras-transformed HaCaT cells resulted in downregulation of MMP-13 and KGFR expression. These results provide evidence, that KGF does not promote progression of cutaneous SCC, but rather suppresses the malignant phenotype of cutaneous SCC cells by regulating the expression of several genes differentially expressed in SCC cells, as compared to normal keratinocytes.
Conflict of interest statement
Figures


Similar articles
-
Expression and roles of keratinocyte growth factor and its receptor in esophageal cancer cells.Int J Oncol. 2007 Oct;31(4):721-8. Int J Oncol. 2007. PMID: 17786302
-
Effects of keratinocyte growth factor on the proliferation and radiation survival of human squamous cell carcinoma cell lines in vitro and in vivo.Int J Radiat Oncol Biol Phys. 1998 Jan 1;40(1):177-87. doi: 10.1016/s0360-3016(97)00561-0. Int J Radiat Oncol Biol Phys. 1998. PMID: 9422575
-
Keratinocyte growth factor induces matrix metalloproteinase-9 expression and correlates with venous invasion in pancreatic cancer.Int J Oncol. 2012 Apr;40(4):1040-8. doi: 10.3892/ijo.2011.1280. Epub 2011 Dec 6. Int J Oncol. 2012. PMID: 22159401 Free PMC article.
-
Potential dual role of KGF/KGFR as a target option in novel therapeutic strategies for the treatment of cancers and mucosal damages.Expert Opin Ther Targets. 2012 Apr;16(4):377-93. doi: 10.1517/14728222.2012.671813. Epub 2012 Mar 25. Expert Opin Ther Targets. 2012. PMID: 22443411 Review.
-
Keratinocyte growth factor expression and activity in cancer: implications for use in patients with solid tumors.J Natl Cancer Inst. 2006 Jun 21;98(12):812-24. doi: 10.1093/jnci/djj228. J Natl Cancer Inst. 2006. PMID: 16788155 Review.
Cited by
-
Complement Factor D Is a Novel Biomarker and Putative Therapeutic Target in Cutaneous Squamous Cell Carcinoma.Cancers (Basel). 2022 Jan 8;14(2):305. doi: 10.3390/cancers14020305. Cancers (Basel). 2022. PMID: 35053469 Free PMC article.
-
RhoGDI1 interacts with PHLDA2, suppresses the proliferation, migration, and invasion of trophoblast cells, and participates in the pathogenesis of preeclampsia.Hum Cell. 2022 Sep;35(5):1440-1452. doi: 10.1007/s13577-022-00746-w. Epub 2022 Jul 16. Hum Cell. 2022. PMID: 35841528
-
Transcriptionally inducible Pleckstrin homology-like domain, family A, member 1, attenuates ErbB receptor activity by inhibiting receptor oligomerization.J Biol Chem. 2018 Feb 9;293(6):2206-2218. doi: 10.1074/jbc.M117.778399. Epub 2017 Dec 12. J Biol Chem. 2018. PMID: 29233889 Free PMC article.
-
Gene Expression Signatures Point to a Male Sex-Specific Lung Mesenchymal Cell PDGF Receptor Signaling Defect in Infants Developing Bronchopulmonary Dysplasia.Sci Rep. 2018 Nov 20;8(1):17070. doi: 10.1038/s41598-018-35256-z. Sci Rep. 2018. PMID: 30459472 Free PMC article.
-
Identification of potential immune-related circRNA-miRNA-mRNA regulatory network in cutaneous squamous cell carcinoma.Am J Cancer Res. 2021 Oct 15;11(10):4826-4843. eCollection 2021. Am J Cancer Res. 2021. PMID: 34765295 Free PMC article.
References
-
- Finch PW, Rubin JS, Miki T, Ron D, Aaronson SA. Human KGF is FGF-related with properties of a paracrine effector of epithelial cell growth. Science. 1989;245:752–755. - PubMed
-
- Jameson J, Ugarte K, Chen N, Yachi P, Fuchs E, et al. A role for skin γδT cells in wound repair. Science. 2002;296:747–749. - PubMed
-
- Andreadis ST, Hamoen KE, Yarmush ML, Morgan JR. Keratinocyte growth factor induces hyperproliferation and delays differentiation in a skin equivalent model system. FASEB J. 2001;15:898–906. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

