Sox9 directs hypertrophic maturation and blocks osteoblast differentiation of growth plate chondrocytes
- PMID: 22421045
- PMCID: PMC3306603
- DOI: 10.1016/j.devcel.2011.12.024
Sox9 directs hypertrophic maturation and blocks osteoblast differentiation of growth plate chondrocytes
Abstract
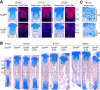
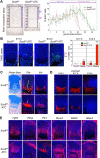
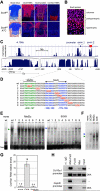
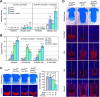
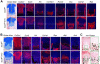
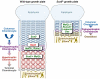
The transcription factor Sox9 is necessary for early chondrogenesis, but its subsequent roles in the cartilage growth plate, a highly specialized structure that drives skeletal growth and endochondral ossification, remain unclear. Using a doxycycline-inducible Cre transgene and Sox9 conditional null alleles in the mouse, we show that Sox9 is required to maintain chondrocyte columnar proliferation and generate cell hypertrophy, two key features of functional growth plates. Sox9 keeps Runx2 expression and β-catenin signaling in check and thereby inhibits not only progression from proliferation to prehypertrophy, but also subsequent acquisition of an osteoblastic phenotype. Sox9 protein outlives Sox9 RNA in upper hypertrophic chondrocytes, where it contributes with Mef2c to directly activate the major marker of these cells, Col10a1. These findings thus reveal that Sox9 remains a central determinant of the lineage fate and multistep differentiation program of growth plate chondrocytes and thereby illuminate our understanding of key molecular mechanisms underlying skeletogenesis.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
SOX9 keeps growth plates and articular cartilage healthy by inhibiting chondrocyte dedifferentiation/osteoblastic redifferentiation.Proc Natl Acad Sci U S A. 2021 Feb 23;118(8):e2019152118. doi: 10.1073/pnas.2019152118. Proc Natl Acad Sci U S A. 2021. PMID: 33597301 Free PMC article.
-
SOX9 governs differentiation stage-specific gene expression in growth plate chondrocytes via direct concomitant transactivation and repression.PLoS Genet. 2011 Nov;7(11):e1002356. doi: 10.1371/journal.pgen.1002356. Epub 2011 Nov 3. PLoS Genet. 2011. PMID: 22072985 Free PMC article.
-
SOX9 is a major negative regulator of cartilage vascularization, bone marrow formation and endochondral ossification.Development. 2010 Mar;137(6):901-11. doi: 10.1242/dev.045203. Development. 2010. PMID: 20179096
-
A pathway to bone: signaling molecules and transcription factors involved in chondrocyte development and maturation.Development. 2015 Mar 1;142(5):817-31. doi: 10.1242/dev.105536. Development. 2015. PMID: 25715393 Free PMC article. Review.
-
Transcriptional networks controlling chondrocyte proliferation and differentiation during endochondral ossification.Pediatr Nephrol. 2010 Apr;25(4):625-31. doi: 10.1007/s00467-009-1368-6. Epub 2009 Dec 1. Pediatr Nephrol. 2010. PMID: 19949815 Review.
Cited by
-
Serum NT/CT SIRT1 ratio reflects early osteoarthritis and chondrosenescence.Ann Rheum Dis. 2020 Oct;79(10):1370-1380. doi: 10.1136/annrheumdis-2020-217072. Epub 2020 Jul 14. Ann Rheum Dis. 2020. PMID: 32665267 Free PMC article.
-
SOX9 gene shows association with adolescent idiopathic scoliosis predisposition in Northwest Indians.Eur J Med Res. 2024 Jan 20;29(1):66. doi: 10.1186/s40001-024-01635-8. Eur J Med Res. 2024. PMID: 38245767 Free PMC article.
-
Precocious chondrocyte differentiation disrupts skeletal growth in Kabuki syndrome mice.JCI Insight. 2019 Oct 17;4(20):e129380. doi: 10.1172/jci.insight.129380. JCI Insight. 2019. PMID: 31557133 Free PMC article.
-
Bone Fracture Acute Phase Response-A Unifying Theory of Fracture Repair: Clinical and Scientific Implications.Clin Rev Bone Miner Metab. 2018;16(4):142-158. doi: 10.1007/s12018-018-9256-x. Epub 2018 Dec 29. Clin Rev Bone Miner Metab. 2018. PMID: 30930699 Free PMC article.
-
Epidermal growth factor signalling pathway in endochondral ossification: an evidence-based narrative review.Ann Med. 2022 Dec;54(1):37-50. doi: 10.1080/07853890.2021.2015798. Ann Med. 2022. PMID: 34955078 Free PMC article. Review.
References
-
- Akiyama H. Control of chondrogenesis by the transcription factor Sox9. Mod. Rheumatol. 2008;18:213–219. - PubMed
-
- Arnold MA, Kim Y, Czubryt MP, Phan D, McAnally J, Qi X, Shelton JM, Richardson JA, Bassel-Duby R, Olson EN. MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev. Cell. 2007;12:377–389. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

