Targeted cancer therapy: giving histone deacetylase inhibitors all they need to succeed
- PMID: 22416777
- PMCID: PMC3341130
- DOI: 10.4155/fmc.12.3
Targeted cancer therapy: giving histone deacetylase inhibitors all they need to succeed
Erratum in
- Future Med Chem. 2012 Jun;4(10):1369-70
Abstract
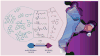
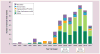
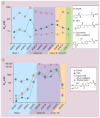
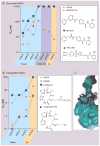
Histone deacetylase inhibitors (HDACis) have now emerged as a powerful new class of small-molecule therapeutics acting through the regulation of the acetylation states of histone proteins (a form of epigenetic modulation) and other non-histone protein targets. Over 490 clinical trials have been initiated in the last 10 years, culminating in the approval of two structurally distinct HDACis - SAHA (vorinostat, Zolinza™) and FK228 (romidepsin, Istodax™). However, the current HDACis have serious limitations, including ineffectively low concentrations in solid tumors and cardiac toxicity, which is hindering their progress in the clinic. Herein, we review the primary paradigms being pursued to overcome these hindrances, including HDAC isoform selectivity, localized administration, and targeting cap groups to achieve selective tissue and cell type distribution.
Figures







Similar articles
-
New and emerging HDAC inhibitors for cancer treatment.J Clin Invest. 2014 Jan;124(1):30-9. doi: 10.1172/JCI69738. Epub 2014 Jan 2. J Clin Invest. 2014. PMID: 24382387 Free PMC article. Review.
-
Selective Histone Deacetylase Inhibitors with Anticancer Activity.Curr Top Med Chem. 2016;16(4):415-26. doi: 10.2174/1568026615666150813145629. Curr Top Med Chem. 2016. PMID: 26268343 Review.
-
Rational therapeutic combinations with histone deacetylase inhibitors for the treatment of cancer.Future Oncol. 2011 Feb;7(2):263-83. doi: 10.2217/fon.11.2. Future Oncol. 2011. PMID: 21345145 Free PMC article. Review.
-
The structural requirements of histone deacetylase inhibitors: SAHA analogs modified at the C5 position display dual HDAC6/8 selectivity.Bioorg Med Chem Lett. 2017 Aug 1;27(15):3254-3258. doi: 10.1016/j.bmcl.2017.06.033. Epub 2017 Jun 13. Bioorg Med Chem Lett. 2017. PMID: 28648461 Free PMC article.
-
Natural Product Inhibitors of Histone Deacetylases as New Anticancer Agents.Curr Protein Pept Sci. 2018;19(3):333-340. doi: 10.2174/1389203718666170106101133. Curr Protein Pept Sci. 2018. PMID: 28059044 Review.
Cited by
-
Epigenetic: a molecular link between testicular cancer and environmental exposures.Front Endocrinol (Lausanne). 2012 Nov 29;3:150. doi: 10.3389/fendo.2012.00150. eCollection 2012. Front Endocrinol (Lausanne). 2012. PMID: 23230429 Free PMC article.
-
Implications of the HDAC6-ERK1 feed-forward loop in immunotherapy.J Immunol Sci. 2018;2(3):59-68. doi: 10.29245/2578-3009/2018/3.1143. Epub 2018 Jun 13. J Immunol Sci. 2018. PMID: 30854521 Free PMC article.
-
Hydroxamic Acid-Based Histone Deacetylase (HDAC) Inhibitors Bearing a Pyrazole Scaffold and a Cinnamoyl Linker.Int J Mol Sci. 2019 Feb 21;20(4):945. doi: 10.3390/ijms20040945. Int J Mol Sci. 2019. PMID: 30795625 Free PMC article.
-
Suberoylanilide hydroxamic acid radiosensitizes tumor hypoxic cells in vitro through the oxidation of nitroxyl to nitric oxide.Free Radic Biol Med. 2014 Aug;73:291-8. doi: 10.1016/j.freeradbiomed.2014.05.019. Epub 2014 May 28. Free Radic Biol Med. 2014. PMID: 24880052 Free PMC article.
-
Transcriptional coregulators: fine-tuning metabolism.Cell Metab. 2014 Jul 1;20(1):26-40. doi: 10.1016/j.cmet.2014.03.027. Epub 2014 May 1. Cell Metab. 2014. PMID: 24794975 Free PMC article. Review.
References
-
- Schulz WA, Hoffmann MJ. Transcription factor networks in embryonic stem cells and testicular cancer and the definition of epigenetics. Epigenetics. 2007;2(1):37–42. - PubMed
-
- Jagannathan V, Robinson-Rechavi M. The challenge of modeling nuclear receptor regulatory networks in mammalian cells. Mol Cell Endocrinol. 2011;334(1–2):91–97. - PubMed
Websites
-
- National Cancer Institute. [Accessed 11 October 2011];Topical romidepsin in treating patients with Stage 1 or Stage 2 cutaneous T-cell non-Hodgkin’s lymphoma. http://clinicaltrials.gov/show/NCT00477698 NLM Identifier: NCT00477698.
-
- Shape Pharmaceuticals Inc. [Accessed 11 October 2011];Safety, Pharmacodynamics (PD), Pharmacokinetics (PK) Study of SHP141 in 1A, 1B, or 2A Cutaneous T-Cell Lymphoma (CTCL) http://clinicaltrials.gov/show/NCT01433731 NLM Identifier: NCT01433731.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
