Quantitative analysis of the detergent-insoluble brain proteome in frontotemporal lobar degeneration using SILAC internal standards
- PMID: 22416763
- PMCID: PMC3357000
- DOI: 10.1021/pr2010814
Quantitative analysis of the detergent-insoluble brain proteome in frontotemporal lobar degeneration using SILAC internal standards
Abstract
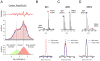
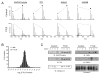
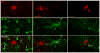
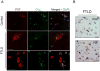
A hallmark of neurodegeneration is the aggregation of disease related proteins that are resistant to detergent extraction. In the major pathological subtype of frontotemporal lobar degeneration (FTLD), modified TAR-DNA binding protein 43 (TDP-43), including phosphorylated, ubiquitinated, and proteolytically cleaved forms, is enriched in detergent-insoluble fractions from post-mortem brain tissue. Additional proteins that accumulate in the detergent-insoluble FTLD brain proteome remain largely unknown. In this study, we used proteins from stable isotope-labeled (SILAC) human embryonic kidney 293 cells (HEK293) as internal standards for peptide quantitation across control and FTLD insoluble brain proteomes. Proteins were identified and quantified by liquid-chromatography coupled with tandem mass spectrometry (LC-MS/MS) and 21 proteins were determined to be enriched in FTLD using SILAC internal standards. In parallel, label-free quantification of only the unlabeled brain derived peptides by spectral counts (SC) and G-test analysis identified additional brain-specific proteins significantly enriched in disease. Several proteins determined to be enriched in FTLD using SILAC internal standards were not considered significant by G-test due to their low total number of SC. However, immunoblotting of FTLD and control samples confirmed enrichment of these proteins, highlighting the utility of SILAC internal standard to quantify low-abundance proteins in brain. Of these, the RNA binding protein PTB-associated splicing factor (PSF) was further characterized because of structural and functional similarities to TDP-43. Full-length PSF and shorter molecular weight fragments, likely resulting from proteolytic cleavage, were enriched in FTLD cases. Immunohistochemical analysis of PSF revealed predominately nuclear localization in control and FTLD brain tissue and was not associated with phosphorylated pathologic TDP-43 neuronal inclusions. However, in a subset of FTLD cases, PSF was aberrantly localized to the cytoplasm of oligodendrocytes. These data raise the possibility that PSF directed RNA processes in oligodendrocytes are altered in neurodegenerative disease.
Figures

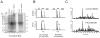
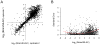

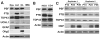
Similar articles
-
Phosphoproteomic analysis reveals site-specific changes in GFAP and NDRG2 phosphorylation in frontotemporal lobar degeneration.J Proteome Res. 2010 Dec 3;9(12):6368-79. doi: 10.1021/pr100666c. Epub 2010 Oct 22. J Proteome Res. 2010. PMID: 20886841 Free PMC article.
-
Aberrant septin 11 is associated with sporadic frontotemporal lobar degeneration.Mol Neurodegener. 2011 Nov 29;6:82. doi: 10.1186/1750-1326-6-82. Mol Neurodegener. 2011. PMID: 22126117 Free PMC article.
-
The RNA-binding motif 45 (RBM45) protein accumulates in inclusion bodies in amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration with TDP-43 inclusions (FTLD-TDP) patients.Acta Neuropathol. 2012 Nov;124(5):717-32. doi: 10.1007/s00401-012-1045-x. Epub 2012 Sep 21. Acta Neuropathol. 2012. PMID: 22993125 Free PMC article.
-
Phosphorylated and cleaved TDP-43 in ALS, FTLD and other neurodegenerative disorders and in cellular models of TDP-43 proteinopathy.Neuropathology. 2010 Apr;30(2):170-81. doi: 10.1111/j.1440-1789.2009.01089.x. Epub 2010 Jan 19. Neuropathology. 2010. PMID: 20102522 Review.
-
Long noncoding RNAs in TDP-43 and FUS/TLS-related frontotemporal lobar degeneration (FTLD).Neurobiol Dis. 2015 Oct;82:445-454. doi: 10.1016/j.nbd.2015.07.011. Epub 2015 Jul 26. Neurobiol Dis. 2015. PMID: 26220395 Review.
Cited by
-
Recent advances in quantitative neuroproteomics.Methods. 2013 Jun 15;61(3):186-218. doi: 10.1016/j.ymeth.2013.04.008. Epub 2013 Apr 25. Methods. 2013. PMID: 23623823 Free PMC article. Review.
-
Selective targeting of the cysteine proteome by thioredoxin and glutathione redox systems.Mol Cell Proteomics. 2013 Nov;12(11):3285-96. doi: 10.1074/mcp.M113.030437. Epub 2013 Aug 14. Mol Cell Proteomics. 2013. PMID: 23946468 Free PMC article.
-
Calcium-responsive transactivator (CREST) protein shares a set of structural and functional traits with other proteins associated with amyotrophic lateral sclerosis.Mol Neurodegener. 2015 Apr 10;10:20. doi: 10.1186/s13024-015-0014-y. Mol Neurodegener. 2015. PMID: 25888396 Free PMC article.
-
Characterization of Detergent Insoluble Proteome in Chronic Traumatic Encephalopathy.J Neuropathol Exp Neurol. 2018 Jan 1;77(1):40-49. doi: 10.1093/jnen/nlx100. J Neuropathol Exp Neurol. 2018. PMID: 29145658 Free PMC article.
-
Detergent Insoluble Proteins and Inclusion Body-Like Structures Immunoreactive for PRKDC/DNA-PK/DNA-PKcs, FTL, NNT, and AIFM1 in the Amygdala of Cognitively Impaired Elderly Persons.J Neuropathol Exp Neurol. 2018 Jan 1;77(1):21-39. doi: 10.1093/jnen/nlx097. J Neuropathol Exp Neurol. 2018. PMID: 29186589 Free PMC article.
References
-
- Taylor JP, Hardy J, Fischbeck KH. Toxic proteins in neurodegenerative disease. Science. 2002;296:1991–5. - PubMed
-
- Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10 (Suppl):S10–7. - PubMed
-
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–3. - PubMed
-
- Mackenzie IR, Rademakers R, Neumann M. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 9:995–1007. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32 AG032848-01A1/AG/NIA NIH HHS/United States
- T32 GM008169/GM/NIGMS NIH HHS/United States
- F32 AG032848-02/AG/NIA NIH HHS/United States
- F32 AG038259/AG/NIA NIH HHS/United States
- F30NS057902/NS/NINDS NIH HHS/United States
- T32 NS007480/NS/NINDS NIH HHS/United States
- P30 NS055077-03/NS/NINDS NIH HHS/United States
- F30 NS057902/NS/NINDS NIH HHS/United States
- P50AG025688/AG/NIA NIH HHS/United States
- P30 NS055077/NS/NINDS NIH HHS/United States
- F32AG032848-02/AG/NIA NIH HHS/United States
- P30NS055077/NS/NINDS NIH HHS/United States
- F32 AG032848/AG/NIA NIH HHS/United States
- P50 AG025688-02/AG/NIA NIH HHS/United States
- P50 AG025688/AG/NIA NIH HHS/United States
- F30 NS057902-02/NS/NINDS NIH HHS/United States
- F32 AG038259-01A2/AG/NIA NIH HHS/United States
- 5T32NS007480/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

