HIV-1 disease progression is associated with bile-salt stimulated lipase (BSSL) gene polymorphism
- PMID: 22412885
- PMCID: PMC3295759
- DOI: 10.1371/journal.pone.0032534
HIV-1 disease progression is associated with bile-salt stimulated lipase (BSSL) gene polymorphism
Abstract
Background: DC-SIGN expressed by dendritic cells captures HIV-1 resulting in trans-infection of CD4(+) T-lymphocytes. However, BSSL (bile-salt stimulated lipase) binding to DC-SIGN interferes with HIV-1 capture. DC-SIGN binding properties of BSSL associate with the polymorphic repeated motif of BSSL exon 11. Furthermore, BSSL binds to HIV-1 co-receptor CXCR4. We hypothesized that BSSL modulates HIV-1 disease progression and emergence of CXCR4 using HIV-1 (X4) variants.
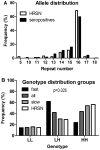
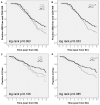
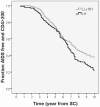
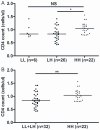
Results: The relation between BSSL genotype and HIV-1 disease progression and emergence of X4 variants was studied using Kaplan Meier and multivariate Cox proportional hazard analysis in a cohort of HIV-1 infected men having sex with men (n = 334, with n = 130 seroconverters). We analyzed the association of BSSL genotype with set-point viral load and CD4 cell count, both pre-infection and post-infection at viral set-point. The number of repeats in BSSL exon 11 were highly variable ranging from 10 to 18 in seropositive individuals and from 5-17 in HRSN with 16 repeats being dominant (>80% carry at least one allele with 16 repeats). We defined 16 to 18 repeats as high (H) and less than 16 repeats as low (L) repeat numbers. Homozygosity for the high (H) repeat number BSSL genotype (HH) correlated with high CD4 cell numbers prior to infection (p = 0.007). In HIV-1 patients, delayed disease progression was linked to the HH BSSL genotype (RH = 0.462 CI = 0.282-0.757, p = 0.002) as was delayed emergence of X4 variants (RH = 0.525, 95% CI = 0.290-0.953, p = 0.034). The LH BSSL genotype, previously found to be associated with enhanced DC-SIGN binding of human milk, was identified to correlate with accelerated disease progression in our cohort of HIV-1 infected MSM (RH = 0.517, 95% CI = 0.328-0.818, p = 0.005).
Conclusion: We identify BSSL as a marker for HIV-1 disease progression and emergence of X4 variants. Additionally, we identified a relation between BSSL genotype and CD4 cell counts prior to infection.
Conflict of interest statement
Figures
Similar articles
-
Binding of human milk to pathogen receptor DC-SIGN varies with bile salt-stimulated lipase (BSSL) gene polymorphism.PLoS One. 2011 Feb 28;6(2):e17316. doi: 10.1371/journal.pone.0017316. PLoS One. 2011. PMID: 21386960 Free PMC article.
-
Bile-salt stimulated lipase polymorphisms do not associate with HCV susceptibility.Virus Res. 2019 Dec;274:197715. doi: 10.1016/j.virusres.2019.197715. Epub 2019 Oct 14. Virus Res. 2019. PMID: 31622635
-
The presence of CXCR4-using HIV-1 prior to start of antiretroviral therapy is an independent predictor of delayed viral suppression.PLoS One. 2013 Oct 1;8(10):e76255. doi: 10.1371/journal.pone.0076255. eCollection 2013. PLoS One. 2013. PMID: 24098454 Free PMC article.
-
Human milk bile salt-stimulated lipase: functional and molecular aspects.J Pediatr. 1994 Nov;125(5 Pt 2):S56-61. doi: 10.1016/s0022-3476(06)80737-7. J Pediatr. 1994. PMID: 7965454 Review.
-
HIV-1 infection in individuals with the CCR5-Delta32/Delta32 genotype: acquisition of syncytium-inducing virus at seroconversion.J Acquir Immune Defic Syndr. 2002 Mar 1;29(3):307-13. doi: 10.1097/00126334-200203010-00013. J Acquir Immune Defic Syndr. 2002. PMID: 11873082 Review.
Cited by
-
Association of TREX1 polymorphism with disease progression in human immunodeficiency virus type-1 (HIV-1) infected patients.Virus Genes. 2023 Dec;59(6):831-835. doi: 10.1007/s11262-023-02032-9. Epub 2023 Sep 20. Virus Genes. 2023. PMID: 37728706
-
Colorectal mucus binds DC-SIGN and inhibits HIV-1 trans-infection of CD4+ T-lymphocytes.PLoS One. 2015 Mar 20;10(3):e0122020. doi: 10.1371/journal.pone.0122020. eCollection 2015. PLoS One. 2015. PMID: 25793526 Free PMC article.
-
Bile salt-stimulated lipase plays an unexpected role in arthritis development in rodents.PLoS One. 2012;7(10):e47006. doi: 10.1371/journal.pone.0047006. Epub 2012 Oct 11. PLoS One. 2012. PMID: 23071697 Free PMC article.
-
Association of interleukin-27 gene polymorphisms with susceptibility to HIV infection and disease progression.J Cell Mol Med. 2019 Apr;23(4):2410-2418. doi: 10.1111/jcmm.14067. Epub 2019 Jan 10. J Cell Mol Med. 2019. PMID: 30632263 Free PMC article.
References
-
- Paxton WA, Kang S. Chemokine receptor allelic polymorphisms: relationships to HIV resistance and disease progression. Semin Immunol. 1998;10:187–194. - PubMed
-
- Smith MW, Dean M, Carrington M, Winkler C, Huttley GA, et al. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC), ALIVE Study. Science. 1997;277:959–965. - PubMed
-
- Winkler C, Modi W, Smith MW, Nelson GW, Wu X, et al. Genetic restriction of AIDS pathogenesis by an SDF-1 chemokine gene variant. ALIVE Study, Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC). Science. 1998;279:389–393. - PubMed
-
- Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

