Transcription goes digital
- PMID: 22410830
- PMCID: PMC3321162
- DOI: 10.1038/embor.2012.31
Transcription goes digital
Abstract
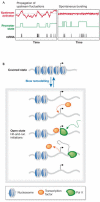
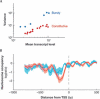
Transcription is a complex process that integrates the state of the cell and its environment to generate adequate responses for cell fitness and survival. Recent microscopy experiments have been able to monitor transcription from single genes in individual cells. These observations have revealed two striking features: transcriptional activity can vary markedly from one cell to another, and is subject to large changes over time, sometimes within minutes. How the chromatin structure, transcription machinery assembly and signalling networks generate such patterns is still unclear. In this review, we present the techniques used to investigate transcription from single genes, introduce quantitative modelling tools, and discuss transcription mechanisms and their implications for gene expression regulation.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
Stochastic models of transcription: from single molecules to single cells.Methods. 2013 Jul 15;62(1):13-25. doi: 10.1016/j.ymeth.2013.03.026. Epub 2013 Apr 1. Methods. 2013. PMID: 23557991
-
On the spontaneous stochastic dynamics of a single gene: complexity of the molecular interplay at the promoter.BMC Syst Biol. 2010 Jan 8;4:2. doi: 10.1186/1752-0509-4-2. BMC Syst Biol. 2010. PMID: 20064204 Free PMC article.
-
Stochastic and delayed stochastic models of gene expression and regulation.Math Biosci. 2010 Jan;223(1):1-11. doi: 10.1016/j.mbs.2009.10.007. Epub 2009 Oct 31. Math Biosci. 2010. PMID: 19883665 Review.
-
Transcription stochasticity of complex gene regulation models.Biophys J. 2012 Sep 19;103(6):1152-61. doi: 10.1016/j.bpj.2012.07.011. Biophys J. 2012. PMID: 22995487 Free PMC article.
-
Message in a nucleus: signaling to the transcriptional machinery.Curr Opin Genet Dev. 2008 Oct;18(5):397-403. doi: 10.1016/j.gde.2008.07.007. Epub 2008 Aug 28. Curr Opin Genet Dev. 2008. PMID: 18678250 Free PMC article. Review.
Cited by
-
Global and targeted approaches to single-cell transcriptome characterization.Brief Funct Genomics. 2018 Jul 1;17(4):209-219. doi: 10.1093/bfgp/elx025. Brief Funct Genomics. 2018. PMID: 29028866 Free PMC article.
-
Live-cell imaging reveals the spatiotemporal organization of endogenous RNA polymerase II phosphorylation at a single gene.Nat Commun. 2021 May 26;12(1):3158. doi: 10.1038/s41467-021-23417-0. Nat Commun. 2021. PMID: 34039974 Free PMC article.
-
DNA residence time is a regulatory factor of transcription repression.Nucleic Acids Res. 2017 Nov 2;45(19):11121-11130. doi: 10.1093/nar/gkx728. Nucleic Acids Res. 2017. PMID: 28977492 Free PMC article.
-
Unified bursting strategies in ectopic and endogenous even-skipped expression patterns.bioRxiv [Preprint]. 2024 Jun 25:2023.02.09.527927. doi: 10.1101/2023.02.09.527927. bioRxiv. 2024. Update in: Elife. 2024 Dec 09;12:RP88671. doi: 10.7554/eLife.88671. PMID: 36798351 Free PMC article. Updated. Preprint.
-
Stochastic transcription in the p53-mediated response to DNA damage is modulated by burst frequency.Mol Syst Biol. 2019 Dec;15(12):e9068. doi: 10.15252/msb.20199068. Mol Syst Biol. 2019. PMID: 31885199 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

