DNA replication fidelity in Escherichia coli: a multi-DNA polymerase affair
- PMID: 22404288
- PMCID: PMC3391330
- DOI: 10.1111/j.1574-6976.2012.00338.x
DNA replication fidelity in Escherichia coli: a multi-DNA polymerase affair
Abstract
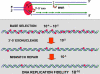
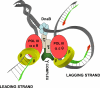
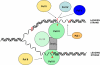
High accuracy (fidelity) of DNA replication is important for cells to preserve the genetic identity and to prevent the accumulation of deleterious mutations. The error rate during DNA replication is as low as 10(-9) to 10(-11) errors per base pair. How this low level is achieved is an issue of major interest. This review is concerned with the mechanisms underlying the fidelity of the chromosomal replication in the model system Escherichia coli by DNA polymerase III holoenzyme, with further emphasis on participation of the other, accessory DNA polymerases, of which E. coli contains four (Pols I, II, IV, and V). Detailed genetic analysis of mutation rates revealed that (1) Pol II has an important role as a back-up proofreader for Pol III, (2) Pols IV and V do not normally contribute significantly to replication fidelity, but can readily do so under conditions of elevated expression, (3) participation of Pols IV and V, in contrast to that of Pol II, is specific to the lagging strand, and (4) Pol I also makes a lagging-strand-specific fidelity contribution, limited, however, to the faithful filling of the Okazaki fragment gaps. The fidelity role of the Pol III τ subunit is also reviewed.
© 2012 Federation of European Microbiological Societies. Published by Blackwell Publishing Ltd. All rights reserved.
Figures

Similar articles
-
Role of accessory DNA polymerases in DNA replication in Escherichia coli: analysis of the dnaX36 mutator mutant.J Bacteriol. 2008 Mar;190(5):1730-42. doi: 10.1128/JB.01463-07. Epub 2007 Dec 21. J Bacteriol. 2008. PMID: 18156258 Free PMC article.
-
DNA polymerase II as a fidelity factor in chromosomal DNA synthesis in Escherichia coli.Mol Microbiol. 2005 Oct;58(1):61-70. doi: 10.1111/j.1365-2958.2005.04805.x. Mol Microbiol. 2005. PMID: 16164549
-
DNA polymerase switching: effects on spontaneous mutagenesis in Escherichia coli.Mol Microbiol. 2009 Jan;71(2):315-31. doi: 10.1111/j.1365-2958.2008.06526.x. Epub 2008 Nov 4. Mol Microbiol. 2009. PMID: 19019142 Free PMC article.
-
A Comprehensive View of Translesion Synthesis in Escherichia coli.Microbiol Mol Biol Rev. 2020 Jun 17;84(3):e00002-20. doi: 10.1128/MMBR.00002-20. Print 2020 Aug 19. Microbiol Mol Biol Rev. 2020. PMID: 32554755 Free PMC article. Review.
-
Insights into the complex levels of regulation imposed on Escherichia coli DNA polymerase V.DNA Repair (Amst). 2016 Aug;44:42-50. doi: 10.1016/j.dnarep.2016.05.005. Epub 2016 May 13. DNA Repair (Amst). 2016. PMID: 27236212 Free PMC article. Review.
Cited by
-
The Rate and Molecular Spectrum of Spontaneous Mutations in the GC-Rich Multichromosome Genome of Burkholderia cenocepacia.Genetics. 2015 Jul;200(3):935-46. doi: 10.1534/genetics.115.176834. Epub 2015 May 12. Genetics. 2015. PMID: 25971664 Free PMC article.
-
Ultraspecific and Amplification-Free Quantification of Mutant DNA by Single-Molecule Kinetic Fingerprinting.J Am Chem Soc. 2018 Sep 19;140(37):11755-11762. doi: 10.1021/jacs.8b06685. Epub 2018 Sep 5. J Am Chem Soc. 2018. PMID: 30125495 Free PMC article.
-
Chromosome structure and DNA replication dynamics during the life cycle of the predatory bacterium Bdellovibrio bacteriovorus.FEMS Microbiol Rev. 2023 Nov 1;47(6):fuad057. doi: 10.1093/femsre/fuad057. FEMS Microbiol Rev. 2023. PMID: 37791401 Free PMC article. Review.
-
Low mutational load and high mutation rate variation in gut commensal bacteria.PLoS Biol. 2020 Mar 10;18(3):e3000617. doi: 10.1371/journal.pbio.3000617. eCollection 2020 Mar. PLoS Biol. 2020. PMID: 32155146 Free PMC article.
-
Novel Escherichia coli active site dnaE alleles with altered base and sugar selectivity.Mol Microbiol. 2021 Sep;116(3):909-925. doi: 10.1111/mmi.14779. Epub 2021 Jul 31. Mol Microbiol. 2021. PMID: 34181784 Free PMC article.
References
-
- Andersson DI, Koskiniemi S, Hughes D. Biological roles of translesion synthesis DNA polymerases in eubacteria. Microbiol. 2010;77:540–548. - PubMed
-
- Banach-Orlowska M, Fijalkowska IJ, Schaaper RM, Jonczyk P. DNA polymerase II as a fidelity factor in chromosomal DNA synthesis in Escherichia coli. Mol Microbiol. 2005;58:61–70. - PubMed
-
- Beard WA, Shock DD, VandeBerg BJ, Wilson SH. Efficiency of correct nucleotide insertion governs DNA polymerase fidelity. J Biol Chem. 2002;277:47393–47398. - PubMed
-
- Becherel OJ, Fuchs RP, Wagner J. Pivotal role of the beta-clamp in translesion DNA synthesis and mutagenesis in E. coli cells. DNA Repair (Amst) 2002;1:703–708. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

