The sub-cellular localization of Sulfolobus DNA replication
- PMID: 22402489
- PMCID: PMC3384333
- DOI: 10.1093/nar/gks217
The sub-cellular localization of Sulfolobus DNA replication
Abstract
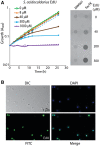
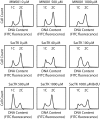
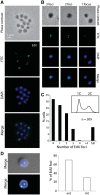
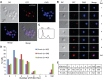
Analyses of the DNA replication-associated proteins of hyperthermophilic archaea have yielded considerable insight into the structure and biochemical function of these evolutionarily conserved factors. However, little is known about the regulation and progression of DNA replication in the context of archaeal cells. In the current work, we describe the generation of strains of Sulfolobus solfataricus and Sulfolobus acidocaldarius that allow the incorporation of nucleoside analogues during DNA replication. We employ this technology, in conjunction with immunolocalization analyses of replisomes, to investigate the sub-cellular localization of nascent DNA and replisomes. Our data reveal a peripheral localization of replisomes in the cell. Furthermore, while the two replication forks emerging from any one of the three replication origins in the Sulfolobus chromosome remain in close proximity, the three origin loci are separated.
Figures

Similar articles
-
The chromosome replication machinery of the archaeon Sulfolobus solfataricus.J Biol Chem. 2006 Jun 2;281(22):15029-32. doi: 10.1074/jbc.R500029200. Epub 2006 Feb 8. J Biol Chem. 2006. PMID: 16467299 Review.
-
Lesion-Induced Mutation in the Hyperthermophilic Archaeon Sulfolobus acidocaldarius and Its Avoidance by the Y-Family DNA Polymerase Dbh.Genetics. 2015 Oct;201(2):513-23. doi: 10.1534/genetics.115.178566. Epub 2015 Jul 29. Genetics. 2015. PMID: 26224736 Free PMC article.
-
Chromosome replication dynamics in the archaeon Sulfolobus acidocaldarius.Proc Natl Acad Sci U S A. 2008 Oct 28;105(43):16737-42. doi: 10.1073/pnas.0806414105. Epub 2008 Oct 15. Proc Natl Acad Sci U S A. 2008. PMID: 18922777 Free PMC article.
-
Extrachromosomal element capture and the evolution of multiple replication origins in archaeal chromosomes.Proc Natl Acad Sci U S A. 2007 Apr 3;104(14):5806-11. doi: 10.1073/pnas.0700206104. Epub 2007 Mar 28. Proc Natl Acad Sci U S A. 2007. PMID: 17392430 Free PMC article.
-
Multiple origins of replication in archaea.Trends Microbiol. 2004 Sep;12(9):399-401. doi: 10.1016/j.tim.2004.07.001. Trends Microbiol. 2004. PMID: 15337158 Review.
Cited by
-
Archaeal imaging: leading the hunt for new discoveries.Mol Biol Cell. 2018 Jul 15;29(13):1675-1681. doi: 10.1091/mbc.E17-10-0603. Mol Biol Cell. 2018. PMID: 30001185 Free PMC article.
-
The use of thermostable fluorescent proteins for live imaging in Sulfolobus acidocaldarius.Front Microbiol. 2024 Sep 9;15:1445186. doi: 10.3389/fmicb.2024.1445186. eCollection 2024. Front Microbiol. 2024. PMID: 39314874 Free PMC article.
-
Establishing Live-Cell Single-Molecule Localization Microscopy Imaging and Single-Particle Tracking in the Archaeon Haloferax volcanii.Front Microbiol. 2020 Nov 20;11:583010. doi: 10.3389/fmicb.2020.583010. eCollection 2020. Front Microbiol. 2020. PMID: 33329447 Free PMC article.
-
Archaeal chromosome biology.J Mol Microbiol Biotechnol. 2014;24(5-6):420-7. doi: 10.1159/000368854. Epub 2015 Feb 17. J Mol Microbiol Biotechnol. 2014. PMID: 25732343 Free PMC article. Review.
-
The cell biology of archaea.Nat Microbiol. 2022 Nov;7(11):1744-1755. doi: 10.1038/s41564-022-01215-8. Epub 2022 Oct 17. Nat Microbiol. 2022. PMID: 36253512 Free PMC article. Review.
References
-
- Duggin IG, Bell SD. The chromosome replication machinery of the archaeon Sulfolobus solfataricus. J. Biol. Chem. 2006;281:15029–15032. - PubMed
-
- Edgell DR, Doolittle WF. Archaea and the origin(s) of DNA replication proteins. Cell. 1997;89:995–998. - PubMed
-
- Robinson NP, Bell SD. Origins of DNA replication in the three domains of life. FEBS J. 2005;272:3757–3766. - PubMed

