Targeted deletion of nrf2 impairs lung development and oxidant injury in neonatal mice
- PMID: 22400915
- PMCID: PMC3423869
- DOI: 10.1089/ars.2011.4288
Targeted deletion of nrf2 impairs lung development and oxidant injury in neonatal mice
Abstract
Aims: Nrf2 is an essential transcription factor for protection against oxidant disorders. However, its role in organ development and neonatal disease has received little attention. Therapeutically administered oxygen has been considered to contribute to bronchopulmonary dysplasia (BPD) in prematurity. The current study was performed to determine Nrf2-mediated molecular events during saccular-to-alveolar lung maturation, and the role of Nrf2 in the pathogenesis of hyperoxic lung injury using newborn Nrf2-deficient (Nrf2(-/-)) and wild-type (Nrf2(+/+)) mice.
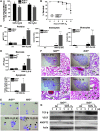
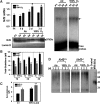
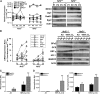
Results: Pulmonary basal expression of cell cycle, redox balance, and lipid/carbohydrate metabolism genes was lower while lymphocyte immunity genes were more highly expressed in Nrf2(-/-) neonates than in Nrf2(+/+) neonates. Hyperoxia-induced phenotypes, including mortality, arrest of saccular-to-alveolar transition, and lung edema, and inflammation accompanying DNA damage and tissue oxidation were significantly more severe in Nrf2(-/-) neonates than in Nrf2(+/+) neonates. During lung injury pathogenesis, Nrf2 orchestrated expression of lung genes involved in organ injury and morphology, cellular growth/proliferation, vasculature development, immune response, and cell-cell interaction. Bioinformatic identification of Nrf2 binding motifs and augmented hyperoxia-induced inflammation in genetically deficient neonates supported Gpx2 and Marco as Nrf2 effectors.
Innovation: This investigation used lung transcriptomics and gene targeted mice to identify novel molecular events during saccular-to-alveolar stage transition and to elucidate Nrf2 downstream mechanisms in protection from hyperoxia-induced injury in neonate mouse lungs.
Conclusion: Nrf2 deficiency augmented lung injury and arrest of alveolarization caused by hyperoxia during the newborn period. Results suggest a therapeutic potential of specific Nrf2 activators for oxidative stress-associated neonatal disorders including BPD.
Figures



Similar articles
-
Sulforaphane enriched transcriptome of lung mitochondrial energy metabolism and provided pulmonary injury protection via Nrf2 in mice.Toxicol Appl Pharmacol. 2019 Feb 1;364:29-44. doi: 10.1016/j.taap.2018.12.004. Epub 2018 Dec 5. Toxicol Appl Pharmacol. 2019. PMID: 30529165 Free PMC article.
-
Transcriptional responses of neonatal mouse lung to hyperoxia by Nrf2 status.Cytokine. 2014 Jan;65(1):4-9. doi: 10.1016/j.cyto.2013.09.021. Epub 2013 Oct 17. Cytokine. 2014. PMID: 24139870 Free PMC article.
-
Preconditioning the immature lung with enhanced Nrf2 activity protects against oxidant-induced hypoalveolarization in mice.Sci Rep. 2020 Nov 4;10(1):19034. doi: 10.1038/s41598-020-75834-8. Sci Rep. 2020. PMID: 33149211 Free PMC article.
-
Molecular Mechanisms of Hyperoxia-Induced Neonatal Intestinal Injury.Int J Mol Sci. 2023 Feb 22;24(5):4366. doi: 10.3390/ijms24054366. Int J Mol Sci. 2023. PMID: 36901800 Free PMC article. Review.
-
Oxygen toxicity: cellular mechanisms in normobaric hyperoxia.Cell Biol Toxicol. 2023 Feb;39(1):111-143. doi: 10.1007/s10565-022-09773-7. Epub 2022 Sep 16. Cell Biol Toxicol. 2023. PMID: 36112262 Free PMC article. Review.
Cited by
-
NRF2 signaling plays an essential role in cancer progression through the NRF2-GPX2-NOTCH3 axis in head and neck squamous cell carcinoma.Oncogenesis. 2024 Sep 27;13(1):35. doi: 10.1038/s41389-024-00536-z. Oncogenesis. 2024. PMID: 39333079 Free PMC article.
-
SIRT1-Related Signaling Pathways and Their Association With Bronchopulmonary Dysplasia.Front Med (Lausanne). 2021 Feb 22;8:595634. doi: 10.3389/fmed.2021.595634. eCollection 2021. Front Med (Lausanne). 2021. PMID: 33693011 Free PMC article. Review.
-
Genomics, microbiomics, proteomics, and metabolomics in bronchopulmonary dysplasia.Semin Perinatol. 2018 Nov;42(7):425-431. doi: 10.1053/j.semperi.2018.09.004. Epub 2018 Oct 2. Semin Perinatol. 2018. PMID: 30487069 Free PMC article. Review.
-
Age-dependent regulation of SARS-CoV-2 cell entry genes and cell death programs correlates with COVID-19 severity.Sci Adv. 2021 Aug 18;7(34):eabf8609. doi: 10.1126/sciadv.abf8609. Print 2021 Aug. Sci Adv. 2021. PMID: 34407940 Free PMC article.
-
N-acetyl-lysyltyrosylcysteine amide, a novel systems pharmacology agent, reduces bronchopulmonary dysplasia in hyperoxic neonatal rat pups.Free Radic Biol Med. 2021 Apr;166:73-89. doi: 10.1016/j.freeradbiomed.2021.02.006. Epub 2021 Feb 17. Free Radic Biol Med. 2021. PMID: 33607217 Free PMC article.
References
-
- Ahola T. Lapatto R. Raivio KO. Selander B. Stigson L. Jonsson B. Jonsbo F. Esberg G. Stovring S. Kjartansson S. Stiris T. Lossius K. Virkola K. Fellman V. N-acetylcysteine does not prevent bronchopulmonary dysplasia in immature infants: a randomized controlled trial. J Pediatr. 2003;143:713–719. - PubMed
-
- Baraldi E. Filippone M. Chronic lung disease after premature birth. N Engl J Med. 2007;357:1946–1955. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

