Versatile pathway-centric approach based on high-throughput sequencing to anticancer drug discovery
- PMID: 22396588
- PMCID: PMC3311343
- DOI: 10.1073/pnas.1200305109
Versatile pathway-centric approach based on high-throughput sequencing to anticancer drug discovery
Abstract
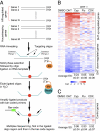
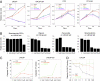
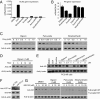
The advent of powerful genomics technologies has uncovered many fundamental aspects of biology, including the mechanisms of cancer; however, it has not been appropriately matched by the development of global approaches to discover new medicines against human diseases. Here we describe a unique high-throughput screening strategy by high-throughput sequencing, referred to as HTS(2), to meet this challenge. This technology enables large-scale and quantitative analysis of gene matrices associated with specific disease phenotypes, therefore allowing screening for small molecules that can specifically intervene with disease-linked gene-expression events. By initially applying this multitarget strategy to the pressing problem of hormone-refractory prostate cancer, which tends to be accelerated by the current antiandrogen therapy, we identify Peruvoside, a cardiac glycoside, which can potently inhibit both androgen-sensitive and -resistant prostate cancer cells without triggering severe cytotoxicity. We further show that, despite transcriptional reprogramming in prostate cancer cells at different disease stages, the compound can effectively block androgen receptor-dependent gene expression by inducing rapid androgen receptor degradation via the proteasome pathway. These findings establish a genomics-based phenotypic screening approach capable of quickly connecting pathways of phenotypic response to the molecular mechanism of drug action, thus offering a unique pathway-centric strategy for drug discovery.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Peruvoside, a Cardiac Glycoside, Induces Primitive Myeloid Leukemia Cell Death.Molecules. 2016 Apr 22;21(4):534. doi: 10.3390/molecules21040534. Molecules. 2016. PMID: 27110755 Free PMC article.
-
Chemical genomics reveals inhibition of breast cancer lung metastasis by Ponatinib via c-Jun.Protein Cell. 2019 Mar;10(3):161-177. doi: 10.1007/s13238-018-0533-8. Epub 2018 Apr 17. Protein Cell. 2019. PMID: 29667003 Free PMC article.
-
Application of high-throughput, molecular-targeted screening to anticancer drug discovery.Curr Top Med Chem. 2002 Mar;2(3):229-46. doi: 10.2174/1568026023394317. Curr Top Med Chem. 2002. PMID: 11944818 Review.
-
Functionalised cyclopentadienyl zirconium compounds as potential anticancer drugs.Dalton Trans. 2008 Oct 21;(39):5293-5. doi: 10.1039/b812244j. Epub 2008 Aug 18. Dalton Trans. 2008. PMID: 18827935
-
A chemical genetics approach for the discovery of apoptosis inducers: from phenotypic cell based HTS assay and structure-activity relationship studies, to identification of potential anticancer agents and molecular targets.Curr Med Chem. 2006;13(22):2627-44. doi: 10.2174/092986706778201521. Curr Med Chem. 2006. PMID: 17017915 Review.
Cited by
-
Large-scale loss-of-function perturbations reveal a comprehensive epigenetic regulatory network in breast cancer.Cancer Biol Med. 2023 Dec 7;21(1):83-103. doi: 10.20892/j.issn.2095-3941.2023.0276. Cancer Biol Med. 2023. PMID: 38062748 Free PMC article.
-
PHDs-seq: a large-scale phenotypic screening method for drug discovery through parallel multi-readout quantification.Cell Regen. 2023 Jun 2;12(1):22. doi: 10.1186/s13619-023-00164-9. Cell Regen. 2023. PMID: 37264282 Free PMC article.
-
Hypoxic Stress-Dependent Regulation of Na,K-ATPase in Ischemic Heart Disease.Int J Mol Sci. 2023 Apr 26;24(9):7855. doi: 10.3390/ijms24097855. Int J Mol Sci. 2023. PMID: 37175562 Free PMC article. Review.
-
Drug Repurposing: A New Hope in Drug Discovery for Prostate Cancer.ACS Omega. 2022 Dec 29;8(1):56-73. doi: 10.1021/acsomega.2c05821. eCollection 2023 Jan 10. ACS Omega. 2022. PMID: 36643505 Free PMC article. Review.
-
Integrating network pharmacology analysis and pharmacodynamic evaluation for exploring the active components and molecular mechanism of moutan seed coat extract to improve cognitive impairment.Front Pharmacol. 2022 Aug 12;13:952876. doi: 10.3389/fphar.2022.952876. eCollection 2022. Front Pharmacol. 2022. PMID: 36034803 Free PMC article.
References
-
- Flordellis CS, Manolis AS, Paris H, Karabinis A. Rethinking target discovery in polygenic diseases. Curr Top Med Chem. 2006;6:1791–1798. - PubMed
-
- Swinney DC, Anthony J. How were new medicines discovered? Nat Rev Drug Discov. 2011;10:507–519. - PubMed
-
- Imming P, Sinning C, Meyer A. Drugs, their targets and the nature and number of drug targets. Nat Rev Drug Discov. 2006;5:821–834. - PubMed
-
- Paul SM, et al. How to improve R&D productivity: The pharmaceutical industry's grand challenge. Nat Rev Drug Discov. 2010;9:203–214. - PubMed
-
- Stegmaier K, et al. Gene expression-based high-throughput screening (GE-HTS) and application to leukemia differentiation. Nat Genet. 2004;36:257–263. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

