Immunogenicity of a lentiviral-based DNA vaccine driven by the 5'LTR of the naturally attenuated caprine arthritis encephalitis virus (CAEV) in mice and macaques
- PMID: 22387218
- PMCID: PMC3341415
- DOI: 10.1016/j.vaccine.2012.02.050
Immunogenicity of a lentiviral-based DNA vaccine driven by the 5'LTR of the naturally attenuated caprine arthritis encephalitis virus (CAEV) in mice and macaques
Abstract
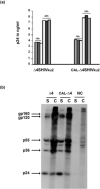
Increasing the safety and the efficacy of existing HIV vaccines is one of the strategies that could help to promote the development of a vaccine for human use. We developed a HIV DNA vaccine (Δ4-SHIVKU2) that has been shown to induce potent polyfunctional HIV-specific T cell responses following a single dose immunization of mice and macaques. Δ4-SHIVKU2 also induced protection when immunized macaques were challenged with homologous pathogenic viruses. In the present study, our aim was to examine whether a chimeric HIV DNA vaccine (CAL-Δ4-SHIVKU2) whose genome is driven by the LTR of the goat lentivirus, caprine arthritis encephalitis (CAEV) expresses efficiently the vaccine antigens and induces potent immune responses in animal models for HIV vaccine. Data of radioimmunoprecipitation assays clearly show that this chimeric genome drives efficient expression of all HIV antigens in the construct. In addition, evaluation of the p24 Gag protein in the supernatant of HEK-293-T cells transfected in parallel with Δ4-SHIVKU2 and CAL-Δ4-SHIVKU2 showed no difference suggesting that these two LTRs are inducing equally the expression of the viral genes. Immunization of mice and macaques using our single dose immunization regimen resulted in induction of similar IFN-γ ELISPOT responses in Δ4-SHIVKU2- and CAL-Δ4-SHIVKU2-treated mice. Similar profiles of T cell responses were also detected both in mice and macaques when multiparametric flow cytometry analyses were performed. Since CAEV LTR is not dependent of Tat to drive viral gene expression and is not functional for integration with HIV integrase, this new vector increases the safety and efficacy of our vaccine vectors and vaccination strategy.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures

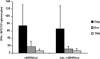


Similar articles
-
A novel non-integrative single-cycle chimeric HIV lentivector DNA vaccine.Vaccine. 2015 May 5;33(19):2273-2282. doi: 10.1016/j.vaccine.2015.03.021. Epub 2015 Mar 28. Vaccine. 2015. PMID: 25825333
-
A noninfectious simian/human immunodeficiency virus DNA vaccine that protects macaques against AIDS.J Virol. 2005 Mar;79(6):3419-28. doi: 10.1128/JVI.79.6.3419-3428.2005. J Virol. 2005. PMID: 15731236 Free PMC article.
-
Characterization of T-cell responses in macaques immunized with a single dose of HIV DNA vaccine.J Virol. 2010 Feb;84(3):1243-53. doi: 10.1128/JVI.01846-09. Epub 2009 Nov 18. J Virol. 2010. PMID: 19923181 Free PMC article.
-
New gene-based approaches for an AIDS vaccine.Comp Immunol Microbiol Infect Dis. 2003 Oct;26(5-6):357-72. doi: 10.1016/S0147-9571(03)00020-1. Comp Immunol Microbiol Infect Dis. 2003. PMID: 12818622 Review.
-
Prime-boost strategies in DNA vaccines.Methods Mol Med. 2006;127:171-97. doi: 10.1385/1-59745-168-1:171. Methods Mol Med. 2006. PMID: 16988455 Review.
Cited by
-
Cowpox Helped Against Smallpox; Will the Goat Lentivirus (Caprine Arthritis Encephalitis Virus) Help Against HIV-1?AIDS Res Hum Retroviruses. 2015 Jun;31(6):577-8. doi: 10.1089/aid.2015.0010. Epub 2015 Apr 6. AIDS Res Hum Retroviruses. 2015. PMID: 25844817 Free PMC article. No abstract available.
-
Long-term central and effector SHIV-specific memory T cell responses elicited after a single immunization with a novel lentivector DNA vaccine.PLoS One. 2014 Oct 22;9(10):e110883. doi: 10.1371/journal.pone.0110883. eCollection 2014. PLoS One. 2014. PMID: 25337803 Free PMC article.
-
Immunization against small ruminant lentiviruses.Viruses. 2013 Aug 2;5(8):1948-63. doi: 10.3390/v5081948. Viruses. 2013. PMID: 23917352 Free PMC article. Review.
-
Tenth International Foamy Virus Conference 2014--achievements and perspectives.Viruses. 2015 Mar 31;7(4):1651-66. doi: 10.3390/v7041651. Viruses. 2015. PMID: 25835535 Free PMC article.
-
Comparative Analysis of Tat-Dependent and Tat-Deficient Natural Lentiviruses.Vet Sci. 2015 Sep 29;2(4):293-348. doi: 10.3390/vetsci2040293. Vet Sci. 2015. PMID: 29061947 Free PMC article. Review.
References
-
- Migueles SA, Laborico AC, Shupert WL, Sabbaghian MS, Rabin R, Hallahan CW, et al. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat Immunol. 2002 Nov;3(11):1061–8. - PubMed
-
- Casartelli N, Di Matteo G, Argentini C, Cancrini C, Bernardi S, Castelli G, et al. Structural defects and variations in the HIV-1 nef gene from rapid, slow and non-progressor children. Aids. 2003 Jun 13;17(9):1291–301. - PubMed
-
- Saksena NK, Ge YC, Wang B, Xiang SH, Dwyer DE, Randle C, et al. An HIV-1 infected long-term non-progressor (LTNP): molecular analysis of HIV-1 strains in the vpr and nef genes. Ann Acad Med Singapore. 1996 Nov;25(6):848–54. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

