Mosquitoes LTR retrotransposons: a deeper view into the genomic sequence of Culex quinquefasciatus
- PMID: 22383973
- PMCID: PMC3286476
- DOI: 10.1371/journal.pone.0030770
Mosquitoes LTR retrotransposons: a deeper view into the genomic sequence of Culex quinquefasciatus
Abstract
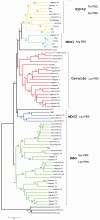
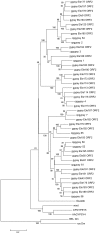
A set of 67 novel LTR-retrotransposon has been identified by in silico analyses of the Culex quinquefasciatus genome using the LTR_STRUC program. The phylogenetic analysis shows that 29 novel and putatively functional LTR-retrotransposons detected belong to the Ty3/gypsy group. Our results demonstrate that, by considering only families containing potentially autonomous LTR-retrotransposons, they account for about 1% of the genome of C. quinquefasciatus. In previous studies it has been estimated that 29% of the genome of C. quinquefasciatus is occupied by mobile genetic elements.The potential role of retrotransposon insertions strictly associated with host genes is described and discussed along with the possible origin of a retrotransposon with peculiar Primer Binding Site region. Finally, we report the presence of a group of 38 retrotransposons, carrying tandem repeated sequences but lacking coding potential, and apparently lacking "master copy" elements from which they could have originated. The features of the repetitive sequences found in these non-autonomous LTR retrotransposons are described, and their possible role discussed.These results integrate the existing data on the genomics of an important virus-borne disease vector.
Conflict of interest statement
Figures

Similar articles
-
Identification of novel LTR retrotransposons in the genome of Aedes aegypti.Gene. 2009 Jul 1;440(1-2):42-9. doi: 10.1016/j.gene.2009.03.021. Epub 2009 Apr 9. Gene. 2009. PMID: 19362135
-
CsRn1, a novel active retrotransposon in a parasitic trematode, Clonorchis sinensis, discloses a new phylogenetic clade of Ty3/gypsy-like LTR retrotransposons.Mol Biol Evol. 2001 Aug;18(8):1474-83. doi: 10.1093/oxfordjournals.molbev.a003933. Mol Biol Evol. 2001. PMID: 11470838
-
Structural and evolutionary analyses of the Ty3/gypsy group of LTR retrotransposons in the genome of Anopheles gambiae.Mol Biol Evol. 2005 Jan;22(1):29-39. doi: 10.1093/molbev/msh251. Epub 2004 Sep 8. Mol Biol Evol. 2005. PMID: 15356275
-
The structure and retrotransposition mechanism of LTR-retrotransposons in the asexual yeast Candida albicans.Virulence. 2014 Aug 15;5(6):655-64. doi: 10.4161/viru.32180. Epub 2014 Aug 7. Virulence. 2014. PMID: 25101670 Free PMC article. Review.
-
Mechanisms of LTR-Retroelement Transposition: Lessons from Drosophila melanogaster.Viruses. 2017 Apr 16;9(4):81. doi: 10.3390/v9040081. Viruses. 2017. PMID: 28420154 Free PMC article. Review.
Cited by
-
LTRsift: a graphical user interface for semi-automatic classification and postprocessing of de novo detected LTR retrotransposons.Mob DNA. 2012 Nov 7;3(1):18. doi: 10.1186/1759-8753-3-18. Mob DNA. 2012. PMID: 23131050 Free PMC article.
-
The Chironomus tentans genome sequence and the organization of the Balbiani ring genes.BMC Genomics. 2014 Sep 27;15(1):819. doi: 10.1186/1471-2164-15-819. BMC Genomics. 2014. PMID: 25261295 Free PMC article.
-
Mosquito genomes are frequently invaded by transposable elements through horizontal transfer.PLoS Genet. 2020 Nov 30;16(11):e1008946. doi: 10.1371/journal.pgen.1008946. eCollection 2020 Nov. PLoS Genet. 2020. PMID: 33253164 Free PMC article.
-
Diverse Defenses: A Perspective Comparing Dipteran Piwi-piRNA Pathways.Cells. 2020 Sep 27;9(10):2180. doi: 10.3390/cells9102180. Cells. 2020. PMID: 32992598 Free PMC article. Review.
-
Transposable element variants and their potential adaptive impact in urban populations of the malaria vector Anopheles coluzzii.Genome Res. 2022 Jan;32(1):189-202. doi: 10.1101/gr.275761.121. Epub 2021 Dec 29. Genome Res. 2022. PMID: 34965939 Free PMC article.
References
-
- Kidwell MG, Lisch DR. Transposable elements as source of genomic variation. In: Craig N, Craigie R, Gellert M, Lambowitz A, editors. Mobile DNA II. American Society for Microbiology Press; 2002. pp. 59–90.
-
- Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–982. - PubMed
-
- Kapitonov VV, Jurka J. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat Rev Genet. 2008;9:411–412; author reply 414. - PubMed
-
- Finnegan DJ. Transposable elements. Curr Opin Genet Dev. 1992;2:861–867. - PubMed
-
- Deragon JM, Casacuberta JM, Panaud O. Plant transposable elements. Genome Dyn. 2008;4:69–82. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

