Role of the ubiquitin-proteasome system in nervous system function and disease: using C. elegans as a dissecting tool
- PMID: 22382927
- PMCID: PMC11115168
- DOI: 10.1007/s00018-012-0946-0
Role of the ubiquitin-proteasome system in nervous system function and disease: using C. elegans as a dissecting tool
Abstract
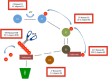
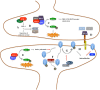
In addition to its central roles in protein quality control, regulation of cell cycle, intracellular signaling, DNA damage response and transcription regulation, the ubiquitin-proteasome system (UPS) plays specific roles in the nervous system, where it contributes to precise connectivity through development, and later assures functionality by regulating a wide spectrum of neuron-specific cellular processes. Aberrations in this system have been implicated in the etiology of neurodevelopmental and neurodegenerative diseases. In this review, we provide an updated view on the UPS and highlight recent findings concerning its role in normal and diseased nervous systems. We discuss the advantages of the model organism Caenorhabditis elegans as a tool to unravel the major unsolved questions concerning this biochemical pathway and its involvement in nervous system function and dysfunction, and expose the new possibilities, using state-of-the-art techniques, to assess UPS function using this model system.
Figures

Similar articles
-
The ubiquitin proteasome system in Caenorhabditis elegans and its regulation.Redox Biol. 2014 Jan 18;2:333-47. doi: 10.1016/j.redox.2014.01.007. eCollection 2014. Redox Biol. 2014. PMID: 24563851 Free PMC article. Review.
-
Failure of ubiquitin proteasome system: risk for neurodegenerative diseases.Neurodegener Dis. 2014;14(4):161-75. doi: 10.1159/000367694. Epub 2014 Nov 20. Neurodegener Dis. 2014. PMID: 25413678 Review.
-
The role of the ubiquitin proteasome system in synapse remodeling and neurodegenerative diseases.Bioessays. 2008 Nov;30(11-12):1075-83. doi: 10.1002/bies.20843. Bioessays. 2008. PMID: 18937340 Free PMC article. Review.
-
Fluorescent Tools for In Vivo Studies on the Ubiquitin-Proteasome System.Methods Mol Biol. 2016;1449:215-22. doi: 10.1007/978-1-4939-3756-1_12. Methods Mol Biol. 2016. PMID: 27613038
-
Tissue-Specific Impact of Autophagy Genes on the Ubiquitin-Proteasome System in C. elegans.Cells. 2020 Aug 8;9(8):1858. doi: 10.3390/cells9081858. Cells. 2020. PMID: 32784405 Free PMC article.
Cited by
-
PRE-1 Revealed Previous Unknown Introgression Events in Eurasian Boars during the Middle Pleistocene.Genome Biol Evol. 2020 Oct 1;12(10):1751-1764. doi: 10.1093/gbe/evaa142. Genome Biol Evol. 2020. PMID: 33151306 Free PMC article.
-
An E2-ubiquitin thioester-driven approach to identify substrates modified with ubiquitin and ubiquitin-like molecules.Nat Commun. 2018 Nov 14;9(1):4776. doi: 10.1038/s41467-018-07251-5. Nat Commun. 2018. PMID: 30429481 Free PMC article.
-
Dominant negative effect of polyglutamine expansion perturbs normal function of ataxin-3 in neuronal cells.Hum Mol Genet. 2015 Jan 1;24(1):100-17. doi: 10.1093/hmg/ddu422. Epub 2014 Aug 20. Hum Mol Genet. 2015. PMID: 25143392 Free PMC article.
-
Associations between Huwe1 and autophagy in rat cerebral neuron oxygen‑glucose deprivation and reperfusion injury.Mol Med Rep. 2020 Dec;22(6):5083-5094. doi: 10.3892/mmr.2020.11611. Epub 2020 Oct 19. Mol Med Rep. 2020. PMID: 33173969 Free PMC article.
-
Identification of shared genes and pathways: a comparative study of multiple sclerosis susceptibility, severity and response to interferon beta treatment.PLoS One. 2013;8(2):e57655. doi: 10.1371/journal.pone.0057655. Epub 2013 Feb 28. PLoS One. 2013. PMID: 23469041 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

