Glycyl-tRNA synthetase specifically binds to the poliovirus IRES to activate translation initiation
- PMID: 22373920
- PMCID: PMC3384309
- DOI: 10.1093/nar/gks182
Glycyl-tRNA synthetase specifically binds to the poliovirus IRES to activate translation initiation
Abstract
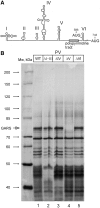
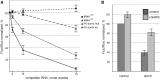
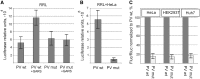
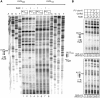
Adaptation to the host cell environment to efficiently take-over the host cell's machinery is crucial in particular for small RNA viruses like picornaviruses that come with only small RNA genomes and replicate exclusively in the cytosol. Their Internal Ribosome Entry Site (IRES) elements are specific RNA structures that facilitate the 5' end-independent internal initiation of translation both under normal conditions and when the cap-dependent host protein synthesis is shut-down in infected cells. A longstanding issue is which host factors play a major role in this internal initiation. Here, we show that the functionally most important domain V of the poliovirus IRES uses tRNA(Gly) anticodon stem-loop mimicry to recruit glycyl-tRNA synthetase (GARS) to the apical part of domain V, adjacent to the binding site of the key initiation factor eIF4G. The binding of GARS promotes the accommodation of the initiation region of the IRES in the mRNA binding site of the ribosome, thereby greatly enhancing the activity of the IRES at the step of the 48S initiation complex formation. Moonlighting functions of GARS that may be additionally needed for other events of the virus-host cell interaction are discussed.
Figures


Similar articles
-
[Glycyl-tRNA Synthetase as a Potential Universal Regulator of Translation Initiation at IRES-I].Mol Biol (Mosk). 2018 Jan-Feb;52(1):10-18. doi: 10.7868/S0026898418010020. Mol Biol (Mosk). 2018. PMID: 29512630 Russian.
-
[Model of the Complex of the Human Glycyl-tRNA Synthetase Anticodon-Binding Domain with IRES I Fragment].Mol Biol (Mosk). 2018 Jan-Feb;52(1):112-119. Mol Biol (Mosk). 2018. PMID: 29512643 Russian.
-
Impaired binding of standard initiation factors eIF3b, eIF4G and eIF4B to domain V of the live-attenuated coxsackievirus B3 Sabin3-like IRES--alternatives for 5'UTR-related cardiovirulence mechanisms.Diagn Pathol. 2013 Sep 24;8:161. doi: 10.1186/1746-1596-8-161. Diagn Pathol. 2013. PMID: 24063684 Free PMC article.
-
Elusive Trans-Acting Factors Which Operate with Type I (Poliovirus-like) IRES Elements.Int J Mol Sci. 2022 Dec 7;23(24):15497. doi: 10.3390/ijms232415497. Int J Mol Sci. 2022. PMID: 36555135 Free PMC article. Review.
-
An atypical IRES within the 5' UTR of a dicistrovirus genome.Virus Res. 2009 Feb;139(2):157-65. doi: 10.1016/j.virusres.2008.07.017. Epub 2008 Sep 11. Virus Res. 2009. PMID: 18755228 Review.
Cited by
-
Associations between Neurological Diseases and Mutations in the Human Glycyl-tRNA Synthetase.Biochemistry (Mosc). 2021 Jan;86(Suppl 1):S12-S23. doi: 10.1134/S0006297921140029. Biochemistry (Mosc). 2021. PMID: 33827397 Free PMC article. Review.
-
Stimulation of the Internal Ribosome Entry Site (IRES)-Dependent Translation of Enterovirus 71 by DDX3X RNA Helicase and Viral 2A and 3C Proteases.Front Microbiol. 2018 Jun 19;9:1324. doi: 10.3389/fmicb.2018.01324. eCollection 2018. Front Microbiol. 2018. PMID: 29971060 Free PMC article.
-
Host-like RNA Elements Regulate Virus Translation.Viruses. 2024 Mar 20;16(3):468. doi: 10.3390/v16030468. Viruses. 2024. PMID: 38543832 Free PMC article. Review.
-
Lipidome is lipids regulator in gastrointestinal tract and it is a life collar in COVID-19: A review.World J Gastroenterol. 2021 Jan 7;27(1):37-54. doi: 10.3748/wjg.v27.i1.37. World J Gastroenterol. 2021. PMID: 33505149 Free PMC article. Review.
-
Dissemination of Internal Ribosomal Entry Sites (IRES) Between Viruses by Horizontal Gene Transfer.Viruses. 2020 Jun 4;12(6):612. doi: 10.3390/v12060612. Viruses. 2020. PMID: 32512856 Free PMC article. Review.
References
-
- Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. - PubMed
-
- Niepmann M. Internal translation initiation of picornaviruses and hepatitis C virus. Biochim. Biophys. Acta. 2009;1789:529–541. - PubMed
-
- Balvay L, Soto Rifo R, Ricci EP, Decimo D, Ohlmann T. Structural and functional diversity of viral IRESes. Biochim. Biophys. Acta. 2009;1789:542–557. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

