A distinct lineage of influenza A virus from bats
- PMID: 22371588
- PMCID: PMC3306675
- DOI: 10.1073/pnas.1116200109
A distinct lineage of influenza A virus from bats
Abstract
Influenza A virus reservoirs in animals have provided novel genetic elements leading to the emergence of global pandemics in humans. Most influenza A viruses circulate in waterfowl, but those that infect mammalian hosts are thought to pose the greatest risk for zoonotic spread to humans and the generation of pandemic or panzootic viruses. We have identified an influenza A virus from little yellow-shouldered bats captured at two locations in Guatemala. It is significantly divergent from known influenza A viruses. The HA of the bat virus was estimated to have diverged at roughly the same time as the known subtypes of HA and was designated as H17. The neuraminidase (NA) gene is highly divergent from all known influenza NAs, and the internal genes from the bat virus diverged from those of known influenza A viruses before the estimated divergence of the known influenza A internal gene lineages. Attempts to propagate this virus in cell cultures and chicken embryos were unsuccessful, suggesting distinct requirements compared with known influenza viruses. Despite its divergence from known influenza A viruses, the bat virus is compatible for genetic exchange with human influenza viruses in human cells, suggesting the potential capability for reassortment and contributions to new pandemic or panzootic influenza A viruses.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


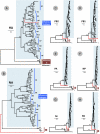
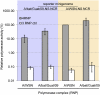
Similar articles
-
Hemagglutinin homologue from H17N10 bat influenza virus exhibits divergent receptor-binding and pH-dependent fusion activities.Proc Natl Acad Sci U S A. 2013 Jan 22;110(4):1458-63. doi: 10.1073/pnas.1218509110. Epub 2013 Jan 7. Proc Natl Acad Sci U S A. 2013. PMID: 23297216 Free PMC article.
-
Crystal structures of two subtype N10 neuraminidase-like proteins from bat influenza A viruses reveal a diverged putative active site.Proc Natl Acad Sci U S A. 2012 Nov 13;109(46):18903-8. doi: 10.1073/pnas.1212579109. Epub 2012 Sep 24. Proc Natl Acad Sci U S A. 2012. PMID: 23012478 Free PMC article.
-
Unusual influenza A viruses in bats.Viruses. 2014 Sep 17;6(9):3438-49. doi: 10.3390/v6093438. Viruses. 2014. PMID: 25256392 Free PMC article. Review.
-
New world bats harbor diverse influenza A viruses.PLoS Pathog. 2013;9(10):e1003657. doi: 10.1371/journal.ppat.1003657. Epub 2013 Oct 10. PLoS Pathog. 2013. PMID: 24130481 Free PMC article.
-
Chiropteran influenza viruses: flu from bats or a relic from the past?Curr Opin Virol. 2016 Feb;16:114-119. doi: 10.1016/j.coviro.2016.02.003. Epub 2016 Mar 3. Curr Opin Virol. 2016. PMID: 26947779 Review.
Cited by
-
Influenza sequence validation and annotation using VADR.Database (Oxford). 2024 Sep 19;2024:baae091. doi: 10.1093/database/baae091. Database (Oxford). 2024. PMID: 39297389 Free PMC article.
-
Prospects for a sequence-based taxonomy of influenza A virus subtypes.Virus Evol. 2024 Aug 17;10(1):veae064. doi: 10.1093/ve/veae064. eCollection 2024. Virus Evol. 2024. PMID: 39247559 Free PMC article.
-
Monitoring of Astroviruses, Brno-Hantaviruses, Coronaviruses, Influenza Viruses, Bornaviruses, Morbilliviruses, Lyssaviruses and Pestiviruses in Austrian Bats.Viruses. 2024 Jul 31;16(8):1232. doi: 10.3390/v16081232. Viruses. 2024. PMID: 39205206 Free PMC article.
-
Incompatible packaging signals and impaired protein functions hinder reassortment of bat H17N10 or H18N11 segment 7 with human H1N1 influenza A viruses.J Virol. 2024 Sep 17;98(9):e0086424. doi: 10.1128/jvi.00864-24. Epub 2024 Aug 20. J Virol. 2024. PMID: 39162567
-
A broad-spectrum vaccine candidate against H5 viruses bearing different sub-clade 2.3.4.4 HA genes.NPJ Vaccines. 2024 Aug 19;9(1):152. doi: 10.1038/s41541-024-00947-4. NPJ Vaccines. 2024. PMID: 39160189 Free PMC article.
References
-
- Reid AH, Taubenberger JK. The origin of the 1918 pandemic influenza virus: A continuing enigma. J Gen Virol. 2003;84:2285–2292. - PubMed
-
- Marx PA, Apetrei C, Drucker E. AIDS as a zoonosis? Confusion over the origin of the virus and the origin of the epidemics. J Med Primatol. 2004;33:220–226. - PubMed
-
- Gao F, et al. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–441. - PubMed
-
- Li W, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

