Insights into assembly and regulation of centromeric chromatin in Saccharomyces cerevisiae
- PMID: 22366340
- PMCID: PMC6296234
- DOI: 10.1016/j.bbagrm.2012.02.008
Insights into assembly and regulation of centromeric chromatin in Saccharomyces cerevisiae
Abstract
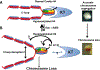
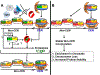
At the core of chromosome segregation is the centromere, which nucleates the assembly of a macromolecular kinetochore (centromere DNA and associated proteins) complex responsible for mediating spindle attachment. Recent advances in centromere research have led to identification of many kinetochore components, such as the centromeric-specific histone H3 variant, CenH3, and its interacting partner, Scm3. Both are essential for chromosome segregation and are evolutionarily conserved from yeast to humans. CenH3 is proposed to be the epigenetic mark that specifies centromeric identity. Molecular mechanisms that regulate the assembly of kinetochores at specific chromosomal sites to mediate chromosome segregation are not fully understood. In this review, we summarize the current literature and discuss results from our laboratory, which show that restricting the localization of budding yeast CenH3, Cse4, to centromeres and balanced stoichiometry between Scm3 and Cse4, contribute to faithful chromosome transmission. We highlight our findings that, similar to other eukaryotic centromeres, budding yeast centromeric histone H4 is hypoacetylated, and we discuss how altered histone acetylation affects chromosome segregation. This article is part of a Special Issue entitled: Chromatin in time and space.
Published by Elsevier B.V.
Figures

Similar articles
-
Misregulation of Scm3p/HJURP causes chromosome instability in Saccharomyces cerevisiae and human cells.PLoS Genet. 2011 Sep;7(9):e1002303. doi: 10.1371/journal.pgen.1002303. Epub 2011 Sep 29. PLoS Genet. 2011. PMID: 21980305 Free PMC article.
-
Scm3 is a centromeric nucleosome assembly factor.J Biol Chem. 2011 Apr 8;286(14):12016-23. doi: 10.1074/jbc.M110.183640. Epub 2011 Feb 12. J Biol Chem. 2011. PMID: 21317428 Free PMC article.
-
Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3.Nature. 2011 Apr 14;472(7342):234-7. doi: 10.1038/nature09854. Epub 2011 Mar 16. Nature. 2011. PMID: 21412236 Free PMC article.
-
Protein kinases in mitotic phosphorylation of budding yeast CENP-A.Curr Genet. 2019 Dec;65(6):1325-1332. doi: 10.1007/s00294-019-00997-5. Epub 2019 May 22. Curr Genet. 2019. PMID: 31119371 Review.
-
Family matters: structural and functional conservation of centromere-associated proteins from yeast to humans.Trends Cell Biol. 2013 Jun;23(6):260-9. doi: 10.1016/j.tcb.2013.01.010. Epub 2013 Mar 5. Trends Cell Biol. 2013. PMID: 23481674 Review.
Cited by
-
Polo kinase Cdc5 associates with centromeres to facilitate the removal of centromeric cohesin during mitosis.Mol Biol Cell. 2016 Jul 15;27(14):2286-300. doi: 10.1091/mbc.E16-01-0004. Epub 2016 May 25. Mol Biol Cell. 2016. PMID: 27226485 Free PMC article.
-
Dbf4-Dependent Kinase (DDK)-Mediated Proteolysis of CENP-A Prevents Mislocalization of CENP-A in Saccharomyces cerevisiae.G3 (Bethesda). 2020 Jun 1;10(6):2057-2068. doi: 10.1534/g3.120.401131. G3 (Bethesda). 2020. PMID: 32295767 Free PMC article.
-
Functional tug of war between kinases, phosphatases, and the Gcn5 acetyltransferase in chromatin and cell cycle checkpoint controls.G3 (Bethesda). 2023 Apr 11;13(4):jkad021. doi: 10.1093/g3journal/jkad021. G3 (Bethesda). 2023. PMID: 36772957 Free PMC article.
-
Phosphorylation of centromeric histone H3 variant regulates chromosome segregation in Saccharomyces cerevisiae.Mol Biol Cell. 2013 Jun;24(12):2034-44. doi: 10.1091/mbc.E12-12-0893. Epub 2013 May 1. Mol Biol Cell. 2013. PMID: 23637466 Free PMC article.
-
Mck1-mediated proteolysis of CENP-A prevents mislocalization of CENP-A for chromosomal stability in Saccharomyces cerevisiae.Genetics. 2024 Sep 4;228(1):iyae108. doi: 10.1093/genetics/iyae108. Genetics. 2024. PMID: 38984710 Free PMC article.
References
-
- Westermann S, Drubin DG, Barnes G, Structures and functions of yeast kineto-chore complexes, Annu. Rev. Biochem 76 (2007) 563–591. - PubMed
-
- Jaspersen SL, Winey M, The budding yeast spindle pole body: structure, duplication, and function, Annu. Rev. Cell Dev. Biol 20 (2004) 1–28. - PubMed
-
- Cheung AL, Deng W, Telomere dysfunction, genome instability and cancer, Front. Biosci 13 (2008) 2075–2090. - PubMed
-
- Nasmyth K, Segregating sister genomes: the molecular biology of chromosome separation, Science 297 (2002) 559–565. - PubMed
-
- Clarke DJ, Bachant J, Kinetochore structure and spindle assembly checkpoint signaling in the budding yeast, Saccharomyces cerevisiae, Front. Biosci 13 (2008) 6787–6819. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

