Specific and sensitive hydrolysis probe-based real-time PCR detection of epidermal growth factor receptor variant III in oral squamous cell carcinoma
- PMID: 22359620
- PMCID: PMC3280998
- DOI: 10.1371/journal.pone.0031723
Specific and sensitive hydrolysis probe-based real-time PCR detection of epidermal growth factor receptor variant III in oral squamous cell carcinoma
Abstract
Background: The tumor-specific EGFR deletion mutant, EGFRvIII, is characterised by ligand-independent constitutive signalling. Tumors expressing EGFRvIII are resistant to current EGFR-targeted therapy. The frequency of EGFRvIII in head and neck squamous cell carcinoma (HNSCC) is disputed and may vary by specific sub-site. The purpose of this study was to measure the occurrence of EGFRvIII mutations in a specific HNSCC subsite, oral squamous cell carcinoma (OSCC), using a novel real-time PCR assay.
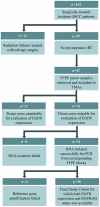
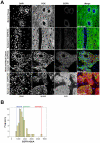
Methodology: Pre-treatment Formalin Fixed Paraffin Embedded (FFPE) cancer specimens from 50 OSCC patients were evaluated for the presence of EGFRvIII using a novel hydrolysis probe-based real-time PCR assay. EGFR protein expression in tumor samples was quantified using fluorescent immunohistochemistry (IHC) and AQUA® technology.
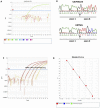
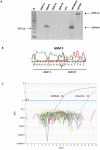
Principal findings: We detected EGFRvIII in a single OSCC patient in our cohort (2%). We confirmed the validity of our detection technique in an independent cohort of glioblastoma patients. We also compared the sensitivity and specificity of our novel real-time EGFRvIII detection assay to conventional RT-PCR and direct sequencing. Our assay can specifically detect EGFRvIII and can discriminate against wild-type EGFR in FFPE tumor samples. AQUAnalysis® revealed that the presence of EGFRvIII transcript is associated with very high EGFR protein expression (98(th) percentile). Contrary to previous reports, only 44% of OSCC over-expressed EGFR in our study.
Conclusion and significance: Our results suggest that the EGFRvIII mutation is rare in OSCC and corroborate previous reports of EGFRvIII expression only in tumors with extreme over-expression of EGFR. We conclude that EGFRvIII-specific therapies may not be ideally suited as first-line treatment in OSCC. Furthermore, highly specific and sensitive methods, such as the real-time RT-PCR assay and AQUAnalysis® described here, will provide accurate assessment of EGFR mutation frequency and EGFR expression, and will facilitate the selection of optimal tailored therapies for OSCC patients.
Conflict of interest statement
Figures
Similar articles
-
Epidermal growth factor receptor variant III in head and neck squamous cell carcinoma is not relevant for targeted therapy and irradiation.Oncotarget. 2017 May 16;8(20):32668-32682. doi: 10.18632/oncotarget.15949. Oncotarget. 2017. PMID: 28427242 Free PMC article.
-
Head and neck squamous cell carcinomas do not express EGFRvIII.Int J Radiat Oncol Biol Phys. 2014 Oct 1;90(2):454-62. doi: 10.1016/j.ijrobp.2014.06.035. Int J Radiat Oncol Biol Phys. 2014. PMID: 25304797
-
Challenges in EGFRvIII detection in head and neck squamous cell carcinoma.PLoS One. 2015 Feb 6;10(2):e0117781. doi: 10.1371/journal.pone.0117781. eCollection 2015. PLoS One. 2015. PMID: 25658924 Free PMC article. Clinical Trial.
-
Epidermal growth factor receptor (EGFR) and squamous cell carcinoma of the skin: molecular bases for EGFR-targeted therapy.Pathol Res Pract. 2011 Jun 15;207(6):337-42. doi: 10.1016/j.prp.2011.03.002. Epub 2011 Apr 29. Pathol Res Pract. 2011. PMID: 21531084 Review.
-
Ribozyme-mediated inhibition of 801-bp deletion-mutant epidermal growth factor receptor mRNA expression in glioblastoma multiforme.Molecules. 2010 Jun 30;15(7):4670-8. doi: 10.3390/molecules15074670. Molecules. 2010. PMID: 20657384 Free PMC article. Review.
Cited by
-
Inhibition of TGF-β and EGF pathway gene expression and migration of oral carcinoma cells by mucosa-associated lymphoid tissue 1.Br J Cancer. 2013 Jul 9;109(1):207-14. doi: 10.1038/bjc.2013.307. Epub 2013 Jun 18. Br J Cancer. 2013. PMID: 23778523 Free PMC article.
-
Phase III randomized, placebo-controlled trial of docetaxel with or without gefitinib in recurrent or metastatic head and neck cancer: an eastern cooperative oncology group trial.J Clin Oncol. 2013 Apr 10;31(11):1405-14. doi: 10.1200/JCO.2012.45.4272. Epub 2013 Mar 4. J Clin Oncol. 2013. PMID: 23460714 Free PMC article. Clinical Trial.
-
Anti-epidermal growth factor receptor therapy in head and neck squamous cell carcinoma: focus on potential molecular mechanisms of drug resistance.Oncologist. 2013;18(7):850-64. doi: 10.1634/theoncologist.2013-0013. Epub 2013 Jul 2. Oncologist. 2013. PMID: 23821327 Free PMC article. Review.
-
Radiolabeled novel mAb 4G1 for immunoSPECT imaging of EGFRvIII expression in preclinical glioblastoma xenografts.Oncotarget. 2017 Jan 24;8(4):6364-6375. doi: 10.18632/oncotarget.14088. Oncotarget. 2017. PMID: 28031526 Free PMC article.
-
Molecular predictors of locoregional and distant metastases in oropharyngeal squamous cell carcinoma.J Otolaryngol Head Neck Surg. 2013 Oct 16;42(1):53. doi: 10.1186/1916-0216-42-53. J Otolaryngol Head Neck Surg. 2013. PMID: 24401183 Free PMC article.
References
-
- Harris TJ, McCormick F. The molecular pathology of cancer. Nature reviews Clinical oncology. 2010;7:251–265. - PubMed
-
- Mitsudomi T, Yatabe Y. Epidermal growth factor receptor in relation to tumor development: EGFR gene and cancer. The FEBS journal. 2010;277:301–308. - PubMed
-
- Lai SY, Johnson FM. Defining the role of the JAK-STAT pathway in head and neck and thoracic malignancies: implications for future therapeutic approaches. Drug resistance updates: reviews and commentaries in antimicrobial and anticancer chemotherapy. 2010;13:67–78. - PubMed
-
- Moscatello DK, Holgado-Madruga M, Godwin AK, Ramirez G, Gunn G, et al. Frequent expression of a mutant epidermal growth factor receptor in multiple human tumors. Cancer research. 1995;55:5536–5539. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

