Composite structural motifs of binding sites for delineating biological functions of proteins
- PMID: 22347478
- PMCID: PMC3275580
- DOI: 10.1371/journal.pone.0031437
Composite structural motifs of binding sites for delineating biological functions of proteins
Abstract
Most biological processes are described as a series of interactions between proteins and other molecules, and interactions are in turn described in terms of atomic structures. To annotate protein functions as sets of interaction states at atomic resolution, and thereby to better understand the relation between protein interactions and biological functions, we conducted exhaustive all-against-all atomic structure comparisons of all known binding sites for ligands including small molecules, proteins and nucleic acids, and identified recurring elementary motifs. By integrating the elementary motifs associated with each subunit, we defined composite motifs that represent context-dependent combinations of elementary motifs. It is demonstrated that function similarity can be better inferred from composite motif similarity compared to the similarity of protein sequences or of individual binding sites. By integrating the composite motifs associated with each protein function, we define meta-composite motifs each of which is regarded as a time-independent diagrammatic representation of a biological process. It is shown that meta-composite motifs provide richer annotations of biological processes than sequence clusters. The present results serve as a basis for bridging atomic structures to higher-order biological phenomena by classification and integration of binding site structures.
Conflict of interest statement
Figures

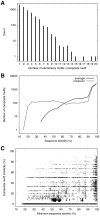
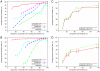
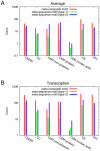
 1) are compared with those with at least one elementary motif (n
1) are compared with those with at least one elementary motif (n 0), the latter are the same as in A. D: Same as C, except that only the UniProt functions of the Biological process category were used.
0), the latter are the same as in A. D: Same as C, except that only the UniProt functions of the Biological process category were used.
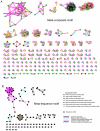
 -trypsin (center) and coagulation factor VII (right) sharing protease inhibitor binding motif (left). D: Cytochrome
-trypsin (center) and coagulation factor VII (right) sharing protease inhibitor binding motif (left). D: Cytochrome  (center) and glycolate oxidase (right) sharing FMN binding motif (left).
(center) and glycolate oxidase (right) sharing FMN binding motif (left).
Similar articles
-
Comprehensive structural classification of ligand-binding motifs in proteins.Structure. 2009 Feb 13;17(2):234-46. doi: 10.1016/j.str.2008.11.009. Structure. 2009. PMID: 19217394
-
Functional structural motifs for protein-ligand, protein-protein, and protein-nucleic acid interactions and their connection to supersecondary structures.Methods Mol Biol. 2013;932:295-315. doi: 10.1007/978-1-62703-065-6_18. Methods Mol Biol. 2013. PMID: 22987360
-
Geometric similarities of protein-protein interfaces at atomic resolution are only observed within homologous families: an exhaustive structural classification study.J Mol Biol. 2010 Jun 11;399(3):526-40. doi: 10.1016/j.jmb.2010.04.028. Epub 2010 Apr 24. J Mol Biol. 2010. PMID: 20417638
-
Structural characteristics of protein binding sites for calcium and lanthanide ions.J Biol Inorg Chem. 2001 Jun;6(5-6):479-89. doi: 10.1007/s007750100214. J Biol Inorg Chem. 2001. PMID: 11472012 Review.
-
Identification of SUMO-binding motifs by NMR.Methods Mol Biol. 2009;497:121-38. doi: 10.1007/978-1-59745-566-4_8. Methods Mol Biol. 2009. PMID: 19107414 Free PMC article. Review.
Cited by
-
A SNARE Protein Identification Method Based on iLearnPlus to Efficiently Solve the Data Imbalance Problem.Front Genet. 2022 Jan 28;12:818841. doi: 10.3389/fgene.2021.818841. eCollection 2021. Front Genet. 2022. PMID: 35154261 Free PMC article.
-
The archiving and dissemination of biological structure data.Curr Opin Struct Biol. 2016 Oct;40:17-22. doi: 10.1016/j.sbi.2016.06.018. Epub 2016 Jul 21. Curr Opin Struct Biol. 2016. PMID: 27450113 Free PMC article. Review.
-
Identifying emerging motif in growing networks.PLoS One. 2014 Jun 17;9(6):e99634. doi: 10.1371/journal.pone.0099634. eCollection 2014. PLoS One. 2014. PMID: 24936978 Free PMC article.
-
GIRAF: a method for fast search and flexible alignment of ligand binding interfaces in proteins at atomic resolution.Biophysics (Nagoya-shi). 2012 May 31;8:79-94. doi: 10.2142/biophysics.8.79. eCollection 2012. Biophysics (Nagoya-shi). 2012. PMID: 27493524 Free PMC article.
-
Structural and functional analysis of multi-interface domains.PLoS One. 2012;7(12):e50821. doi: 10.1371/journal.pone.0050821. Epub 2012 Dec 14. PLoS One. 2012. PMID: 23272073 Free PMC article.
References
-
- Friedberg I. Automated protein function prediction – the genomic challenge. Brief Bioinform. 2006;7:225–242. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

