Coordinating tissue interactions: Notch signaling in cardiac development and disease
- PMID: 22340493
- PMCID: PMC3285259
- DOI: 10.1016/j.devcel.2012.01.014
Coordinating tissue interactions: Notch signaling in cardiac development and disease
Abstract
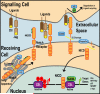
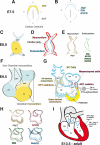
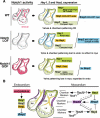
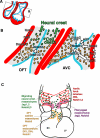
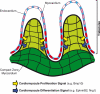
The Notch pathway is a crucial cell-fate regulator in the developing heart. Attention in the past centered on Notch function in cardiomyocytes. However, recent advances demonstrate that region-specific endocardial Notch activity orchestrates the patterning and morphogenesis of cardiac chambers and valves through regulatory interaction with multiple myocardial and neural crest signals. Notch also regulates cardiomyocyte proliferation and differentiation during ventricular chamber development and is required for coronary vessel specification. Here, we review these data and highlight disease connections, including evidence that Notch-Hey-Bmp2 interplay impacts adult heart valve disease and that Notch contributes to cardiac arrhythmia and pre-excitation syndromes.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
Similar articles
-
Notch signaling in cardiac valve development and disease.Birth Defects Res A Clin Mol Teratol. 2011 Jun;91(6):449-59. doi: 10.1002/bdra.20815. Epub 2011 May 11. Birth Defects Res A Clin Mol Teratol. 2011. PMID: 21563298 Review.
-
Endocardial Notch Signaling in Cardiac Development and Disease.Circ Res. 2016 Jan 8;118(1):e1-e18. doi: 10.1161/CIRCRESAHA.115.305350. Epub 2015 Dec 3. Circ Res. 2016. PMID: 26635389 Review.
-
Notch and interacting signalling pathways in cardiac development, disease, and regeneration.Nat Rev Cardiol. 2018 Nov;15(11):685-704. doi: 10.1038/s41569-018-0100-2. Nat Rev Cardiol. 2018. PMID: 30287945 Review.
-
Bmp2 and Notch cooperate to pattern the embryonic endocardium.Development. 2018 Jul 2;145(13):dev163378. doi: 10.1242/dev.163378. Development. 2018. PMID: 29853617
-
Integration of a Notch-dependent mesenchymal gene program and Bmp2-driven cell invasiveness regulates murine cardiac valve formation.J Clin Invest. 2010 Oct;120(10):3493-507. doi: 10.1172/JCI42666. Epub 2010 Sep 20. J Clin Invest. 2010. PMID: 20890042 Free PMC article.
Cited by
-
Delta-Like Ligand 4-Notch Signaling in Macrophage Activation.Arterioscler Thromb Vasc Biol. 2016 Oct;36(10):2038-47. doi: 10.1161/ATVBAHA.116.306926. Epub 2016 Aug 25. Arterioscler Thromb Vasc Biol. 2016. PMID: 27562914 Free PMC article. Review.
-
A review on regulation of DNA methylation during post-myocardial infarction.Front Pharmacol. 2024 Feb 13;15:1267585. doi: 10.3389/fphar.2024.1267585. eCollection 2024. Front Pharmacol. 2024. PMID: 38414735 Free PMC article. Review.
-
Spatiotemporal Gene Coexpression and Regulation in Mouse Cardiomyocytes of Early Cardiac Morphogenesis.J Am Heart Assoc. 2019 Aug 6;8(15):e012941. doi: 10.1161/JAHA.119.012941. Epub 2019 Jul 19. J Am Heart Assoc. 2019. PMID: 31322043 Free PMC article.
-
Angiotensin II Signal Transduction: An Update on Mechanisms of Physiology and Pathophysiology.Physiol Rev. 2018 Jul 1;98(3):1627-1738. doi: 10.1152/physrev.00038.2017. Physiol Rev. 2018. PMID: 29873596 Free PMC article. Review.
-
A personalized, multiomics approach identifies genes involved in cardiac hypertrophy and heart failure.NPJ Syst Biol Appl. 2018 Feb 24;4:12. doi: 10.1038/s41540-018-0046-3. eCollection 2018. NPJ Syst Biol Appl. 2018. PMID: 29507758 Free PMC article.
References
-
- Aanhaanen WT, Brons JF, Dominguez JN, Rana MS, Norden J, Airik R, Wakker V, de Gier-de Vries C, Brown NA, Kispert A, et al. The Tbx2+ primary myocardium of the atrioventricular canal forms the atrioventricular node and the base of the left ventricle. Circ Res. 2009;104:1267–1274. - PubMed
-
- Aanhaanen WT, Moorman AF, Christoffels VM. Origin and development of the atrioventricular myocardial lineage: insight into the development of accessory pathways. Birth Defects Res A Clin Mol Teratol. 2011;91:565–577. - PubMed
-
- Andersson ER, Sandberg R, Lendahl U. Notch signaling: simplicity in design, versatility in function. Development. 2011;138:3593–3612. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

