Regulation of bone-renal mineral and energy metabolism: the PHEX, FGF23, DMP1, MEPE ASARM pathway
- PMID: 22339660
- PMCID: PMC3362997
- DOI: 10.1615/critreveukargeneexpr.v22.i1.50
Regulation of bone-renal mineral and energy metabolism: the PHEX, FGF23, DMP1, MEPE ASARM pathway
Abstract
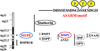
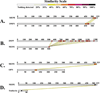
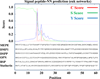
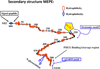
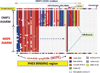
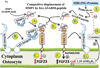
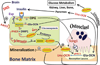
More than 300 million years ago, vertebrates emerged from the vast oceans to conquer gravity and the dry land. With this transition, new adaptations occurred that included ingenious changes in reproduction, waste secretion, and bone physiology. One new innovation, the egg shell, contained an ancestral protein (ovocleidin-116) that likely first appeared with the dinosaurs and was preserved through the theropod lineage in modern birds and reptiles. Ovocleidin-116 is an avian homolog of matrix extracellular phosphoglycoprotein (MEPE) and belongs to a group of proteins called short integrin-binding ligand-interacting glycoproteins (SIBLINGs). These proteins are all localized to a defined region on chromosome 5q in mice and chromosome 4q in humans. A unifying feature of SIBLING proteins is an acidic serine aspartate-rich MEPE-associated motif (ASARM). Recent research has shown that the ASARM motif and the released ASARM peptide have regulatory roles in mineralization (bone and teeth), phosphate regulation, vascularization, soft-tissue calcification, osteoclastogenesis, mechanotransduction, and fat energy metabolism. The MEPE ASARM motif and peptide are physiological substrates for PHEX, a zinc metalloendopeptidase. Defects in PHEX are responsible for X-linked hypophosphatemic rickets (HYP). There is evidence that PHEX interacts with another ASARM motif containing SIBLING protein, dentin matrix protein-1 (DMP1). DMP1 mutations cause bone and renal defects that are identical with the defects caused by a loss of PHEX function. This results in autosomal recessive hypophosphatemic rickets (ARHR). In both HYP and ARHR, increased FGF23 expression plays a major role in the disease and in autosomal dominant hypophosphatemic rickets (ADHR), FGF23 half-life is increased by activating mutations. ASARM peptide administration in vitro and in vivo also induces increased FGF23 expression. FGF23 is a member of the fibroblast growth factor (FGF) family of cytokines, which surfaced 500 million years ago with the boney fish (i.e., teleosts) that do not contain SIBLING proteins. In terrestrial vertebrates, FGF23, like SIBLING proteins, is expressed in the osteocyte. The boney fish, however, are an-osteocytic, so a physiological bone-renal link with FGF23 and the SIBLINGs was cemented when life ventured from the oceans to the land during the Triassic period, approximately 300 million years ago. This link has been revealed by recent research that indicates a competitive displacement of a PHEX-DMP1 interaction by an ASARM peptide that leads to increased FGF23 expression. This review discusses the new discoveries that reveal a novel PHEX, DMP1, MEPE, ASARM peptide, and FGF23 bone-renal pathway. This pathway impacts not only bone formation, bone-renal mineralization, and renal phosphate homeostasis but also energy metabolism. The study of this new pathway is relevant for developing therapies for several diseases: bone-teeth mineral loss disorders, renal osteodystrophy, chronic kidney disease and bone mineralization disorders (CKD-MBD), end-stage renal diseases, ectopic arterial-calcification, cardiovascular disease renal calcification, diabetes, and obesity.
Figures




Similar articles
-
The chicken or the egg: PHEX, FGF23 and SIBLINGs unscrambled.Cell Biochem Funct. 2012 Jul;30(5):355-75. doi: 10.1002/cbf.2841. Epub 2012 May 9. Cell Biochem Funct. 2012. PMID: 22573484 Free PMC article. Review.
-
Degradation of MEPE, DMP1, and release of SIBLING ASARM-peptides (minhibins): ASARM-peptide(s) are directly responsible for defective mineralization in HYP.Endocrinology. 2008 Apr;149(4):1757-72. doi: 10.1210/en.2007-1205. Epub 2007 Dec 27. Endocrinology. 2008. PMID: 18162525 Free PMC article.
-
SPR4-peptide alters bone metabolism of normal and HYP mice.Bone. 2015 Mar;72:23-33. doi: 10.1016/j.bone.2014.11.011. Epub 2014 Nov 22. Bone. 2015. PMID: 25460577 Free PMC article.
-
Phosphorylated acidic serine-aspartate-rich MEPE-associated motif peptide from matrix extracellular phosphoglycoprotein inhibits phosphate regulating gene with homologies to endopeptidases on the X-chromosome enzyme activity.J Endocrinol. 2007 Jan;192(1):261-7. doi: 10.1677/joe.1.07059. J Endocrinol. 2007. PMID: 17210763 Free PMC article.
-
The wrickkened pathways of FGF23, MEPE and PHEX.Crit Rev Oral Biol Med. 2004 Sep 1;15(5):264-81. doi: 10.1177/154411130401500503. Crit Rev Oral Biol Med. 2004. PMID: 15470265 Free PMC article. Review.
Cited by
-
MAF1, a repressor of RNA polymerase III-dependent transcription, regulates bone mass.Elife. 2022 May 25;11:e74740. doi: 10.7554/eLife.74740. Elife. 2022. PMID: 35611941 Free PMC article.
-
Genetic variations for the eggshell crystal structure revealed by genome-wide association study in chickens.BMC Genomics. 2021 Nov 2;22(1):786. doi: 10.1186/s12864-021-08103-1. BMC Genomics. 2021. PMID: 34727889 Free PMC article.
-
The Intricacies of Renal Phosphate Reabsorption-An Overview.Int J Mol Sci. 2024 Apr 25;25(9):4684. doi: 10.3390/ijms25094684. Int J Mol Sci. 2024. PMID: 38731904 Free PMC article. Review.
-
Associations between the levels of sclerostin, phosphate, and fibroblast growth factor-23 and treatment with vitamin D in hemodialysis patients with low intact PTH level.Osteoporos Int. 2015 Mar;26(3):1017-28. doi: 10.1007/s00198-014-2934-8. Epub 2014 Nov 4. Osteoporos Int. 2015. PMID: 25366373
-
Vitamin D attenuates COVID-19 complications via modulation of proinflammatory cytokines, antiviral proteins, and autophagy.Expert Rev Anti Infect Ther. 2022 Feb;20(2):231-241. doi: 10.1080/14787210.2021.1941871. Epub 2021 Jul 15. Expert Rev Anti Infect Ther. 2022. PMID: 34112047 Free PMC article. Review.
References
-
- Burland C. An historic case of rickets: being an account of the medical examination of the remains of Princess Elizabeth, daughter of King Charles I. who died at Carrisbrooke Castle, September 8, 1650. Practitioner. 1918;100:391–395.
-
- TEC J. Princess Elizabeth, second daughter of King Charles 1, and rickets. Pediatrics. 1979;64(2):241.
-
- O’Riordan JL. Rickets in the 17th century. J Bone Miner Res. 2006;21(10):1506–1510. - PubMed
-
- Glisson F, Bate G, Regemorter A. A treatise of the rickets: being a disease common to children. London: Peter Cole, Cornhill Royal Exchange (Bedford Royal Infirmary Medical Library); p. 1651.
-
- Rafter GW. Elmer McCollum and the disappearance of rickets. Perspect Biol Med. 1987;30(4):527–534. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

