Iterative stable alignment and clustering of 2D transmission electron microscope images
- PMID: 22325773
- PMCID: PMC3426367
- DOI: 10.1016/j.str.2011.12.007
Iterative stable alignment and clustering of 2D transmission electron microscope images
Abstract
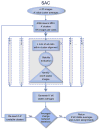
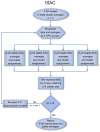
Identification of homogeneous subsets of images in a macromolecular electron microscopy (EM) image data set is a critical step in single-particle analysis. The task is handled by iterative algorithms, whose performance is compromised by the compounded limitations of image alignment and K-means clustering. Here we describe an approach, iterative stable alignment and clustering (ISAC) that, relying on a new clustering method and on the concepts of stability and reproducibility, can extract validated, homogeneous subsets of images. ISAC requires only a small number of simple parameters and, with minimal human intervention, can eliminate bias from two-dimensional image clustering and maximize the quality of group averages that can be used for ab initio three-dimensional structural determination and analysis of macromolecular conformational variability. Repeated testing of the stability and reproducibility of a solution within ISAC eliminates heterogeneous or incorrect classes and introduces critical validation to the process of EM image clustering.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures



Similar articles
-
A Stochastic Hill Climbing Approach for Simultaneous 2D Alignment and Clustering of Cryogenic Electron Microscopy Images.Structure. 2016 Jun 7;24(6):988-96. doi: 10.1016/j.str.2016.04.006. Epub 2016 May 12. Structure. 2016. PMID: 27184214
-
Ab initio structure determination from electron microscopic images of single molecules coexisting in different functional states.Structure. 2010 Jul 14;18(7):777-86. doi: 10.1016/j.str.2010.06.001. Structure. 2010. PMID: 20637414
-
A Fast Image Alignment Approach for 2D Classification of Cryo-EM Images Using Spectral Clustering.Curr Issues Mol Biol. 2021 Oct 18;43(3):1652-1668. doi: 10.3390/cimb43030117. Curr Issues Mol Biol. 2021. PMID: 34698131 Free PMC article.
-
Advances in image processing for single-particle analysis by electron cryomicroscopy and challenges ahead.Curr Opin Struct Biol. 2018 Oct;52:127-145. doi: 10.1016/j.sbi.2018.11.004. Epub 2018 Nov 30. Curr Opin Struct Biol. 2018. PMID: 30509756 Review.
-
[A review of automatic particle recognition in Cryo-EM images].Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 2010 Oct;27(5):1178-82. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 2010. PMID: 21089695 Review. Chinese.
Cited by
-
Structure of mammalian Mediator complex reveals Tail module architecture and interaction with a conserved core.Nat Commun. 2021 Mar 1;12(1):1355. doi: 10.1038/s41467-021-21601-w. Nat Commun. 2021. PMID: 33649303 Free PMC article.
-
Unique double-ring structure of the peroxisomal Pex1/Pex6 ATPase complex revealed by cryo-electron microscopy.Proc Natl Acad Sci U S A. 2015 Jul 28;112(30):E4017-25. doi: 10.1073/pnas.1500257112. Epub 2015 Jul 13. Proc Natl Acad Sci U S A. 2015. PMID: 26170309 Free PMC article.
-
A molecular device for the redox quality control of GroEL/ES substrates.Cell. 2023 Mar 2;186(5):1039-1049.e17. doi: 10.1016/j.cell.2023.01.013. Epub 2023 Feb 9. Cell. 2023. PMID: 36764293 Free PMC article.
-
It takes two transducins to activate the cGMP-phosphodiesterase 6 in retinal rods.Open Biol. 2018 Aug;8(8):180075. doi: 10.1098/rsob.180075. Open Biol. 2018. PMID: 30068566 Free PMC article.
-
Integrative structure and functional anatomy of a nuclear pore complex.Nature. 2018 Mar 22;555(7697):475-482. doi: 10.1038/nature26003. Epub 2018 Mar 14. Nature. 2018. PMID: 29539637 Free PMC article.
References
-
- Armache KJ, Mitterweger S, Meinhart A, Cramer P. Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J Biol Chem. 2005;280:7131–7134. - PubMed
-
- Baldwin PR, Penczek PA. The transform class in SPARX and EMAN2. J Struct Biol. 2007;157:250–261. - PubMed
-
- Burkard RE, Mauro D, Martello S. Assignment Problems. Philadelphia: Society for Industrial and Applied Mathematics; 2009.
-
- Duda RO, Hart PE, Stork DG. Pattern Classification. New York: Wiley; 2001.
-
- Grundel DA, Pardalos PM. Test problem generator for the multidimensional assignment problem. Comput Optim Appl. 2005;30:133–146.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

