A comparison of cataloged variation between International HapMap Consortium and 1000 Genomes Project data
- PMID: 22319179
- PMCID: PMC3277631
- DOI: 10.1136/amiajnl-2011-000652
A comparison of cataloged variation between International HapMap Consortium and 1000 Genomes Project data
Abstract
Background: Since publication of the human genome in 2003, geneticists have been interested in risk variant associations to resolve the etiology of traits and complex diseases. The International HapMap Consortium undertook an effort to catalog all common variation across the genome (variants with a minor allele frequency (MAF) of at least 5% in one or more ethnic groups). HapMap along with advances in genotyping technology led to genome-wide association studies which have identified common variants associated with many traits and diseases. In 2008 the 1000 Genomes Project aimed to sequence 2500 individuals and identify rare variants and 99% of variants with a MAF of <1%.
Methods: To determine whether the 1000 Genomes Project includes all the variants in HapMap, we examined the overlap between single nucleotide polymorphisms (SNPs) genotyped in the two resources using merged phase II/III HapMap data and low coverage pilot data from 1000 Genomes.
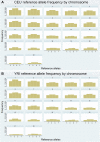
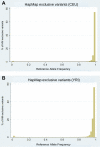
Results: Comparison of the two data sets showed that approximately 72% of HapMap SNPs were also found in 1000 Genomes Project pilot data. After filtering out HapMap variants with a MAF of <5% (separately for each population), 99% of HapMap SNPs were found in 1000 Genomes data.
Conclusions: Not all variants cataloged in HapMap are also cataloged in 1000 Genomes. This could affect decisions about which resource to use for SNP queries, rare variant validation, or imputation. Both the HapMap and 1000 Genomes Project databases are useful resources for human genetics, but it is important to understand the assumptions made and filtering strategies employed by these projects.
Conflict of interest statement
Figures


Similar articles
-
Comparing genetic variants detected in the 1000 genomes project with SNPs determined by the International HapMap Consortium.J Genet. 2015 Dec;94(4):731-40. doi: 10.1007/s12041-015-0588-8. J Genet. 2015. PMID: 26690529
-
Comprehensive evaluation of imputation performance in African Americans.J Hum Genet. 2012 Jul;57(7):411-21. doi: 10.1038/jhg.2012.43. Epub 2012 May 31. J Hum Genet. 2012. PMID: 22648186 Free PMC article.
-
Genotype imputation for African Americans using data from HapMap phase II versus 1000 genomes projects.Genet Epidemiol. 2012 Jul;36(5):508-16. doi: 10.1002/gepi.21647. Epub 2012 May 29. Genet Epidemiol. 2012. PMID: 22644746 Free PMC article.
-
Human genetics and genomics a decade after the release of the draft sequence of the human genome.Hum Genomics. 2011 Oct;5(6):577-622. doi: 10.1186/1479-7364-5-6-577. Hum Genomics. 2011. PMID: 22155605 Free PMC article. Review.
-
[Analysis and application of SNP and haplotype in the human genome].Yi Chuan Xue Bao. 2005 Aug;32(8):879-89. Yi Chuan Xue Bao. 2005. PMID: 16231744 Review. Chinese.
Cited by
-
Distinct Transcript Isoforms of the Atypical Chemokine Receptor 1 (ACKR1)/Duffy Antigen Receptor for Chemokines (DARC) Gene Are Expressed in Lymphoblasts and Altered Isoform Levels Are Associated with Genetic Ancestry and the Duffy-Null Allele.PLoS One. 2015 Oct 16;10(10):e0140098. doi: 10.1371/journal.pone.0140098. eCollection 2015. PLoS One. 2015. PMID: 26473357 Free PMC article.
-
The genetic basis of quality of life in healthy Swedish women: a candidate gene approach.PLoS One. 2015 Feb 12;10(2):e0118292. doi: 10.1371/journal.pone.0118292. eCollection 2015. PLoS One. 2015. PMID: 25675377 Free PMC article.
-
Association Between the MUC5B Promoter Polymorphism rs35705950 and Idiopathic Pulmonary Fibrosis: A Meta-analysis and Trial Sequential Analysis in Caucasian and Asian Populations.Medicine (Baltimore). 2015 Oct;94(43):e1901. doi: 10.1097/MD.0000000000001901. Medicine (Baltimore). 2015. PMID: 26512610 Free PMC article.
-
Comparing genetic variants detected in the 1000 genomes project with SNPs determined by the International HapMap Consortium.J Genet. 2015 Dec;94(4):731-40. doi: 10.1007/s12041-015-0588-8. J Genet. 2015. PMID: 26690529
-
Self-reported race/ethnicity in the age of genomic research: its potential impact on understanding health disparities.Hum Genomics. 2015 Jan 7;9(1):1. doi: 10.1186/s40246-014-0023-x. Hum Genomics. 2015. PMID: 25563503 Free PMC article. Review.
References
-
- Donis-Keller H, Green P, Helms C, et al. A genetic linkage map of the human genome. Cell 1987;51:319–37 - PubMed
-
- Weissenbach J, Gyapay G, Dib C, et al. A second-generation linkage map of the human genome. Nature 1992;359:794–801 - PubMed
-
- Patterson K. 1000 genomes: a world of variation. Circ Res 2011;108:534–6 - PubMed

