DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes
- PMID: 22314321
- PMCID: PMC3459314
- DOI: 10.1038/nature10909
DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes
Abstract
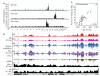
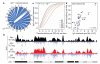
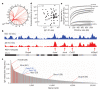
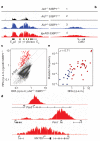
Recurrent chromosomal translocations underlie both haematopoietic and solid tumours. Their origin has been ascribed to selection of random rearrangements, targeted DNA damage, or frequent nuclear interactions between translocation partners; however, the relative contribution of each of these elements has not been measured directly or on a large scale. Here we examine the role of nuclear architecture and frequency of DNA damage in the genesis of chromosomal translocations by measuring these parameters simultaneously in cultured mouse B lymphocytes. In the absence of recurrent DNA damage, translocations between Igh or Myc and all other genes are directly related to their contact frequency. Conversely, translocations associated with recurrent site-directed DNA damage are proportional to the rate of DNA break formation, as measured by replication protein A accumulation at the site of damage. Thus, non-targeted rearrangements reflect nuclear organization whereas DNA break formation governs the location and frequency of recurrent translocations, including those driving B-cell malignancies.
Figures
Similar articles
-
Mechanisms promoting translocations in editing and switching peripheral B cells.Nature. 2009 Jul 9;460(7252):231-6. doi: 10.1038/nature08159. Nature. 2009. PMID: 19587764 Free PMC article.
-
Translocation capture sequencing: a method for high throughput mapping of chromosomal rearrangements.J Immunol Methods. 2012 Jan 31;375(1-2):176-81. doi: 10.1016/j.jim.2011.10.007. Epub 2011 Oct 18. J Immunol Methods. 2012. PMID: 22033343 Free PMC article.
-
Role of genomic instability and p53 in AID-induced c-myc-Igh translocations.Nature. 2006 Mar 2;440(7080):105-9. doi: 10.1038/nature04495. Epub 2006 Jan 8. Nature. 2006. PMID: 16400328 Free PMC article.
-
The origin of recurrent translocations in recombining lymphocytes: a balance between break frequency and nuclear proximity.Curr Opin Cell Biol. 2013 Jun;25(3):365-71. doi: 10.1016/j.ceb.2013.02.007. Epub 2013 Mar 13. Curr Opin Cell Biol. 2013. PMID: 23478218 Free PMC article. Review.
-
AID and Igh switch region-Myc chromosomal translocations.DNA Repair (Amst). 2006 Sep 8;5(9-10):1259-64. doi: 10.1016/j.dnarep.2006.05.019. Epub 2006 Jun 19. DNA Repair (Amst). 2006. PMID: 16784901 Review.
Cited by
-
Spatial re-organization of myogenic regulatory sequences temporally controls gene expression.Nucleic Acids Res. 2015 Feb 27;43(4):2008-21. doi: 10.1093/nar/gkv046. Epub 2015 Feb 4. Nucleic Acids Res. 2015. PMID: 25653159 Free PMC article.
-
Oncogenic Myc translocations are independent of chromosomal location and orientation of the immunoglobulin heavy chain locus.Proc Natl Acad Sci U S A. 2012 Aug 21;109(34):13728-32. doi: 10.1073/pnas.1202882109. Epub 2012 Aug 6. Proc Natl Acad Sci U S A. 2012. PMID: 22869734 Free PMC article.
-
Concurrent V(D)J recombination and DNA end instability increase interchromosomal trans-rearrangements in ATM-deficient thymocytes.Nucleic Acids Res. 2013 Apr;41(8):4535-48. doi: 10.1093/nar/gkt154. Epub 2013 Mar 6. Nucleic Acids Res. 2013. PMID: 23470994 Free PMC article.
-
RNA Exosome Regulates AID DNA Mutator Activity in the B Cell Genome.Adv Immunol. 2015;127:257-308. doi: 10.1016/bs.ai.2015.04.002. Epub 2015 May 14. Adv Immunol. 2015. PMID: 26073986 Free PMC article. Review.
-
Response to Casellas et al.Mol Cell. 2013 Aug 8;51(3):277-8. doi: 10.1016/j.molcel.2013.07.019. Mol Cell. 2013. PMID: 23932710 Free PMC article. No abstract available.
References
-
- Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nature Rev. Cancer. 2007;7:233–245. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

