Calcium/calmodulin inhibition of the Arabidopsis BRASSINOSTEROID-INSENSITIVE 1 receptor kinase provides a possible link between calcium and brassinosteroid signalling
- PMID: 22309147
- PMCID: PMC3316158
- DOI: 10.1042/BJ20111871
Calcium/calmodulin inhibition of the Arabidopsis BRASSINOSTEROID-INSENSITIVE 1 receptor kinase provides a possible link between calcium and brassinosteroid signalling
Abstract
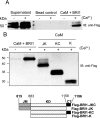
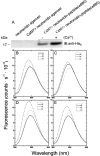
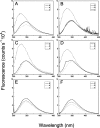
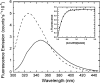
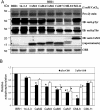
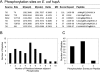
The receptor kinase BRI1 (BRASSINOSTEROID-INSENSITIVE 1) is a key component in BR (brassinosteroid) perception and signal transduction, and has a broad impact on plant growth and development. In the present study, we demonstrate that Arabidopsis CaM (calmodulin) binds to the recombinant cytoplasmic domain of BRI1 in a Ca2+-dependent manner in vitro. In silico analysis predicted binding to Helix E of the BRI1 kinase subdomain VIa and a synthetic peptide based on this sequence interacted with Ca2+/CaM. Co-expression of CaM with the cytoplasmic domain of BRI1 in Escherichia coli strongly reduced autophosphorylation of BRI1, in particular on tyrosine residues, and also reduced the BRI1-mediated transphosphorylation of E. coli proteins on tyrosine, threonine and presumably serine residues. Several isoforms of CaM and CMLs (CaM-like proteins) were more effective (AtCaM6, AtCaM7 and AtCML8, where At is Arabidopsis thaliana) than others (AtCaM2, AtCaM4 and AtCML11) when co-expressed with BRI1 in E. coli. These results establish a novel assay for recombinant BRI1 transphosphorylation activity and collectively uncover a possible new link between Ca2+ and BR signalling.
Figures


Similar articles
-
14-3-3 proteins contribute to leaf and root development via brassinosteroid insensitive 1 in Arabidopsis thaliana.Genes Genomics. 2020 Mar;42(3):347-354. doi: 10.1007/s13258-019-00909-4. Epub 2020 Jan 4. Genes Genomics. 2020. PMID: 31902106
-
Crystal structures of the phosphorylated BRI1 kinase domain and implications for brassinosteroid signal initiation.Plant J. 2014 Apr;78(1):31-43. doi: 10.1111/tpj.12445. Epub 2014 Mar 12. Plant J. 2014. PMID: 24461462 Free PMC article.
-
Ligand perception, activation, and early signaling of plant steroid receptor brassinosteroid insensitive 1.J Integr Plant Biol. 2013 Dec;55(12):1198-211. doi: 10.1111/jipb.12081. Epub 2013 Sep 9. J Integr Plant Biol. 2013. PMID: 23718739 Review.
-
The brassinosteroid receptor BRI1 can generate cGMP enabling cGMP-dependent downstream signaling.Plant J. 2017 Aug;91(4):590-600. doi: 10.1111/tpj.13589. Epub 2017 Jun 12. Plant J. 2017. PMID: 28482142
-
BAK1 directly regulates brassinosteroid perception and BRI1 activation.J Integr Plant Biol. 2013 Dec;55(12):1264-70. doi: 10.1111/jipb.12122. J Integr Plant Biol. 2013. PMID: 24308570 Review.
Cited by
-
Cross Talk Between Cyclic Nucleotides and Calcium Signaling Pathways in Plants-Achievements and Prospects.Front Plant Sci. 2021 Feb 16;12:643560. doi: 10.3389/fpls.2021.643560. eCollection 2021. Front Plant Sci. 2021. PMID: 33664763 Free PMC article. Review.
-
Roles of Brassinosteroids in Mitigating Heat Stress Damage in Cereal Crops.Int J Mol Sci. 2021 Mar 8;22(5):2706. doi: 10.3390/ijms22052706. Int J Mol Sci. 2021. PMID: 33800127 Free PMC article. Review.
-
A simple in vitro method to measure autophosphorylation of protein kinases.Plant Methods. 2013 Jun 26;9(1):22. doi: 10.1186/1746-4811-9-22. Plant Methods. 2013. PMID: 23803530 Free PMC article.
-
Towards Understanding Plant Calcium Signaling through Calmodulin-Like Proteins: A Biochemical and Structural Perspective.Int J Mol Sci. 2018 Apr 30;19(5):1331. doi: 10.3390/ijms19051331. Int J Mol Sci. 2018. PMID: 29710867 Free PMC article. Review.
-
Teaching an old hormone new tricks: cytosolic Ca2+ elevation involvement in plant brassinosteroid signal transduction cascades.Plant Physiol. 2013 Oct;163(2):555-65. doi: 10.1104/pp.112.213371. Epub 2013 Jul 12. Plant Physiol. 2013. PMID: 23852441 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

