Identification of a minimal region of the HIV-1 5'-leader required for RNA dimerization, NC binding, and packaging
- PMID: 22306406
- PMCID: PMC3296369
- DOI: 10.1016/j.jmb.2012.01.033
Identification of a minimal region of the HIV-1 5'-leader required for RNA dimerization, NC binding, and packaging
Abstract
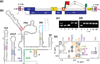
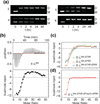
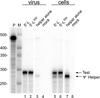
Assembly of human immunodeficiency virus type 1 (HIV-1) particles is initiated in the cytoplasm by the formation of a ribonucleoprotein complex comprising the dimeric RNA genome and a small number of viral Gag polyproteins. Genomes are recognized by the nucleocapsid (NC) domains of Gag, which interact with packaging elements believed to be located primarily within the 5'-leader (5'-L) of the viral RNA. Recent studies revealed that the native 5'-L exists as an equilibrium of two conformers, one in which dimer-promoting residues and NC binding sites are sequestered and packaging is attenuated, and one in which these sites are exposed and packaging is promoted. To identify the elements within the dimeric 5'-L that are important for packaging, we generated HIV-1 5'-L RNAs containing mutations and deletions designed to eliminate substructures without perturbing the overall structure of the leader and examined effects of the mutations on RNA dimerization, NC binding, and packaging. Our findings identify a 159-residue RNA packaging signal that possesses dimerization and NC binding properties similar to those of the intact 5'-L and contains elements required for efficient RNA packaging.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures
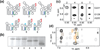

Similar articles
-
5'-Cap sequestration is an essential determinant of HIV-1 genome packaging.Proc Natl Acad Sci U S A. 2021 Sep 14;118(37):e2112475118. doi: 10.1073/pnas.2112475118. Proc Natl Acad Sci U S A. 2021. PMID: 34493679 Free PMC article.
-
Unpaired Guanosines in the 5' Untranslated Region of HIV-1 RNA Act Synergistically To Mediate Genome Packaging.J Virol. 2020 Oct 14;94(21):e00439-20. doi: 10.1128/JVI.00439-20. Print 2020 Oct 14. J Virol. 2020. PMID: 32796062 Free PMC article.
-
Role of distal zinc finger of nucleocapsid protein in genomic RNA dimerization of human immunodeficiency virus type 1; no role for the palindrome crowning the R-U5 hairpin.Virology. 2001 Mar 1;281(1):109-16. doi: 10.1006/viro.2000.0778. Virology. 2001. PMID: 11222101
-
Structural determinants and mechanism of HIV-1 genome packaging.J Mol Biol. 2011 Jul 22;410(4):609-33. doi: 10.1016/j.jmb.2011.04.029. J Mol Biol. 2011. PMID: 21762803 Free PMC article. Review.
-
Retroviral Gag protein-RNA interactions: Implications for specific genomic RNA packaging and virion assembly.Semin Cell Dev Biol. 2019 Feb;86:129-139. doi: 10.1016/j.semcdb.2018.03.015. Epub 2018 Apr 1. Semin Cell Dev Biol. 2019. PMID: 29580971 Free PMC article. Review.
Cited by
-
Sequences within both the 5' UTR and Gag are required for optimal in vivo packaging and propagation of mouse mammary tumor virus (MMTV) genomic RNA.PLoS One. 2012;7(10):e47088. doi: 10.1371/journal.pone.0047088. Epub 2012 Oct 16. PLoS One. 2012. PMID: 23077548 Free PMC article.
-
Distinct Gag interaction properties of HIV-1 RNA 5' leader conformers reveal a mechanism for dimeric genome selection.RNA. 2023 Feb;29(2):217-227. doi: 10.1261/rna.079347.122. Epub 2022 Nov 16. RNA. 2023. PMID: 36384962 Free PMC article.
-
The biased nucleotide composition of the HIV genome: a constant factor in a highly variable virus.Retrovirology. 2012 Nov 6;9:92. doi: 10.1186/1742-4690-9-92. Retrovirology. 2012. PMID: 23131071 Free PMC article. Review.
-
RNA structure. Structure of the HIV-1 RNA packaging signal.Science. 2015 May 22;348(6237):917-21. doi: 10.1126/science.aaa9266. Science. 2015. PMID: 25999508 Free PMC article.
-
Retroviral PBS-segment sequence and structure: Orchestrating early and late replication events.Retrovirology. 2024 Jun 17;21(1):12. doi: 10.1186/s12977-024-00646-x. Retrovirology. 2024. PMID: 38886829 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

