Annexin A2 is involved in the formation of hepatitis C virus replication complex on the lipid raft
- PMID: 22301157
- PMCID: PMC3318618
- DOI: 10.1128/JVI.06327-11
Annexin A2 is involved in the formation of hepatitis C virus replication complex on the lipid raft
Abstract
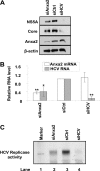
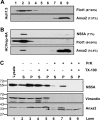
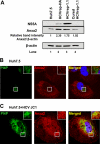
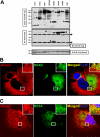
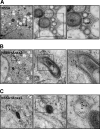
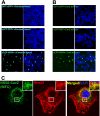
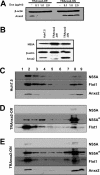
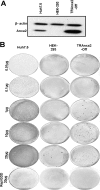
The hepatitis C virus (HCV) RNA replicates in hepatic cells by forming a replication complex on the lipid raft (detergent-resistant membrane [DRM]). Replication complex formation requires various viral nonstructural (NS) proteins as well as host cellular proteins. In our previous study (C. K. Lai, K. S. Jeng, K. Machida, and M. M. Lai, J. Virol. 82:8838-8848, 2008), we found that a cellular protein, annexin A2 (Anxa2), interacts with NS3/NS4A. Since NS3/NS4A is a membranous protein and Anxa2 is known as a lipid raft-associated scaffold protein, we postulate that Anxa2 helps in the formation of the HCV replication complex on the lipid raft. Further studies showed that Anxa2 was localized at the HCV-induced membranous web and interacted with NS4B, NS5A, and NS5B and colocalized with them in the perinuclear region. The silencing of Anxa2 decreased the formation of membranous web-like structures and viral RNA replication. Subcellular fractionation and bimolecular fluorescence complementation analysis revealed that Anxa2 was partially associated with HCV at the lipid raft enriched with phosphatidylinositol-4-phosphate (PI4P) and caveolin-2. Further, the overexpression of Anxa2 in HCV-nonsusceptible HEK293 cells caused the enrichment of HCV NS proteins in the DRM fraction and increased the colony-forming ability of the HCV replicon. Since Anxa2 is known to induce the formation of the lipid raft microdomain, we propose that Anxa2 recruits HCV NS proteins and enriches them on the lipid raft to form the HCV replication complex.
Figures

Similar articles
-
Characterization of interactions between hepatitis C virus NS5B polymerase, annexin A2 and RNA - effects on NS5B catalysis and allosteric inhibition.Virol J. 2017 Dec 11;14(1):236. doi: 10.1186/s12985-017-0904-4. Virol J. 2017. PMID: 29228983 Free PMC article.
-
Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft.J Virol. 2004 Apr;78(7):3480-8. doi: 10.1128/jvi.78.7.3480-3488.2004. J Virol. 2004. PMID: 15016871 Free PMC article.
-
Role of annexin A2 in the production of infectious hepatitis C virus particles.J Virol. 2010 Jun;84(11):5775-89. doi: 10.1128/JVI.02343-09. Epub 2010 Mar 24. J Virol. 2010. PMID: 20335258 Free PMC article.
-
[Suppression of hepatitis C virus (HCV) replication with serine palmitoyltransferase inhibitor].Yakugaku Zasshi. 2010 Feb;130(2):157-61. doi: 10.1248/yakushi.130.157. Yakugaku Zasshi. 2010. PMID: 20118637 Review. Japanese.
-
Hepatitis C virus and host cell lipids: an intimate connection.RNA Biol. 2011 Mar-Apr;8(2):258-69. doi: 10.4161/rna.8.2.15011. Epub 2011 Mar 1. RNA Biol. 2011. PMID: 21593584 Review.
Cited by
-
Autophagy during viral infection - a double-edged sword.Nat Rev Microbiol. 2018 Jun;16(6):341-354. doi: 10.1038/s41579-018-0003-6. Nat Rev Microbiol. 2018. PMID: 29556036 Free PMC article. Review.
-
Characterization of interactions between hepatitis C virus NS5B polymerase, annexin A2 and RNA - effects on NS5B catalysis and allosteric inhibition.Virol J. 2017 Dec 11;14(1):236. doi: 10.1186/s12985-017-0904-4. Virol J. 2017. PMID: 29228983 Free PMC article.
-
Molecular Insights on the Possible Role of Annexin A2 in COVID-19 Pathogenesis and Post-Infection Complications.Int J Mol Sci. 2021 Oct 13;22(20):11028. doi: 10.3390/ijms222011028. Int J Mol Sci. 2021. PMID: 34681689 Free PMC article. Review.
-
Sterol Binding by the Tombusviral Replication Proteins Is Essential for Replication in Yeast and Plants.J Virol. 2017 Mar 13;91(7):e01984-16. doi: 10.1128/JVI.01984-16. Print 2017 Apr 1. J Virol. 2017. PMID: 28100609 Free PMC article.
-
Annexin A2 gene interacting with viral matrix protein to promote bovine ephemeral fever virus release.J Vet Sci. 2020 Mar;21(2):e33. doi: 10.4142/jvs.2020.21.e33. J Vet Sci. 2020. PMID: 32233139 Free PMC article.
References
-
- Aizaki H, Choi KS, Liu M, Li YJ, Lai MM. 2006. Polypyrimidine-tract-binding protein is a component of the HCV RNA replication complex and necessary for RNA synthesis. J. Biomed. Sci. 13:469–480 - PubMed
-
- Aizaki H, Lee KJ, Sung VM, Ishiko H, Lai MM. 2004. Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology 324:450–461 - PubMed
-
- Ayala-Sanmartin J, Henry JP, Pradel LA. 2001. Cholesterol regulates membrane binding and aggregation by annexin 2 at submicromolar Ca(2+) concentration. Biochim. Biophys. Acta 1510:18–28 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

