Assembly and architecture of HIV
- PMID: 22297526
- PMCID: PMC6743068
- DOI: 10.1007/978-1-4614-0980-9_20
Assembly and architecture of HIV
Abstract
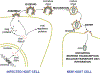
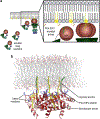
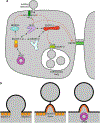
HIV forms spherical, membrane-enveloped, pleomorphic virions, 1,000-1,500 Å in diameter, which contain two copies of its single-stranded, positive-sense RNA genome. Virus particles initially bud from host cells in a noninfectious or immature form, in which the genome is further encapsulated inside a spherical protein shell composed of around 2,500 copies of the virally encoded Gag polyprotein. The Gag molecules are radially arranged, adherent to the inner leaflet of the viral membrane, and closely associated as a hexagonal, paracrystalline lattice. Gag comprises three major structural domains called MA, CA, and NC. For immature virions to become infectious, they must undergo a maturation process that is initiated by proteolytic processing of Gag by the viral protease. The new Gag-derived proteins undergo dramatic rearrangements to form the mature virus. The mature MA protein forms a "matrix" layer and remains attached to the viral envelope, NC condenses with the genome, and approximately 1,500 copies of CA assemble into a new cone-shaped protein shell, called the mature capsid, which surrounds the genomic ribonucleoprotein complex. The HIV capsid conforms to the mathematical principles of a fullerene shell, in which the CA subunits form about 250 CA hexamers arrayed on a variably curved hexagonal lattice, which is closed by incorporation of exactly 12 pentamers, seven pentamers at the wide end and five at the narrow end of the cone. This chapter describes our current understanding of HIV's virion architecture and its dynamic transformations: the process of virion assembly as orchestrated by Gag, the architecture of the immature virion, the virus maturation process, and the structure of the mature capsid.
Figures


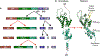



Similar articles
-
RNA and Nucleocapsid Are Dispensable for Mature HIV-1 Capsid Assembly.J Virol. 2015 Oct;89(19):9739-47. doi: 10.1128/JVI.00750-15. Epub 2015 Jul 15. J Virol. 2015. PMID: 26178992 Free PMC article.
-
Identification of a Structural Element in HIV-1 Gag Required for Virus Particle Assembly and Maturation.mBio. 2018 Oct 16;9(5):e01567-18. doi: 10.1128/mBio.01567-18. mBio. 2018. PMID: 30327442 Free PMC article.
-
Structure of the immature retroviral capsid at 8 Å resolution by cryo-electron microscopy.Nature. 2012 Jul 19;487(7407):385-9. doi: 10.1038/nature11169. Nature. 2012. PMID: 22722831
-
The structural biology of HIV assembly.Curr Opin Struct Biol. 2008 Apr;18(2):203-17. doi: 10.1016/j.sbi.2008.02.001. Epub 2008 Apr 9. Curr Opin Struct Biol. 2008. PMID: 18406133 Free PMC article. Review.
-
Exploring HIV-1 Maturation: A New Frontier in Antiviral Development.Viruses. 2024 Sep 6;16(9):1423. doi: 10.3390/v16091423. Viruses. 2024. PMID: 39339899 Free PMC article. Review.
Cited by
-
The Assembly of HTLV-1-How Does It Differ from HIV-1?Viruses. 2024 Sep 27;16(10):1528. doi: 10.3390/v16101528. Viruses. 2024. PMID: 39459862 Free PMC article. Review.
-
Roles of Capsid-Interacting Host Factors in Multimodal Inhibition of HIV-1 by PF74.J Virol. 2016 May 27;90(12):5808-5823. doi: 10.1128/JVI.03116-15. Print 2016 Jun 15. J Virol. 2016. PMID: 27076642 Free PMC article.
-
A protein ballet around the viral genome orchestrated by HIV-1 reverse transcriptase leads to an architectural switch: from nucleocapsid-condensed RNA to Vpr-bridged DNA.Virus Res. 2013 Feb;171(2):287-303. doi: 10.1016/j.virusres.2012.09.008. Epub 2012 Sep 24. Virus Res. 2013. PMID: 23017337 Free PMC article.
-
Crystal structure of an HIV assembly and maturation switch.Elife. 2016 Jul 14;5:e17063. doi: 10.7554/eLife.17063. Elife. 2016. PMID: 27416583 Free PMC article.
-
A temporospatial map that defines specific steps at which critical surfaces in the Gag MA and CA domains act during immature HIV-1 capsid assembly in cells.J Virol. 2014 May;88(10):5718-41. doi: 10.1128/JVI.03609-13. Epub 2014 Mar 12. J Virol. 2014. PMID: 24623418 Free PMC article.
References
-
- Bailey GD, Hyun JK, Mitra AK et al. (2009) Proton-linked dimerization of a retroviral capsid protein initiates capsid assembly. Structure 17:737–748 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

