graphite - a Bioconductor package to convert pathway topology to gene network
- PMID: 22292714
- PMCID: PMC3296647
- DOI: 10.1186/1471-2105-13-20
graphite - a Bioconductor package to convert pathway topology to gene network
Abstract
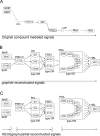
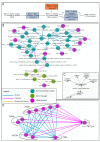
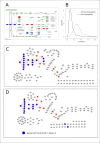
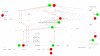
Background: Gene set analysis is moving towards considering pathway topology as a crucial feature. Pathway elements are complex entities such as protein complexes, gene family members and chemical compounds. The conversion of pathway topology to a gene/protein networks (where nodes are a simple element like a gene/protein) is a critical and challenging task that enables topology-based gene set analyses.Unfortunately, currently available R/Bioconductor packages provide pathway networks only from single databases. They do not propagate signals through chemical compounds and do not differentiate between complexes and gene families.
Results: Here we present graphite, a Bioconductor package addressing these issues. Pathway information from four different databases is interpreted following specific biologically-driven rules that allow the reconstruction of gene-gene networks taking into account protein complexes, gene families and sensibly removing chemical compounds from the final graphs. The resulting networks represent a uniform resource for pathway analyses. Indeed, graphite provides easy access to three recently proposed topological methods. The graphite package is available as part of the Bioconductor software suite.
Conclusions: graphite is an innovative package able to gather and make easily available the contents of the four major pathway databases. In the field of topological analysis graphite acts as a provider of biological information by reducing the pathway complexity considering the biological meaning of the pathway elements.
Figures

Similar articles
-
ToPASeq: an R package for topology-based pathway analysis of microarray and RNA-Seq data.BMC Bioinformatics. 2015 Oct 29;16:350. doi: 10.1186/s12859-015-0763-1. BMC Bioinformatics. 2015. PMID: 26514335 Free PMC article.
-
Graphite Web: Web tool for gene set analysis exploiting pathway topology.Nucleic Acids Res. 2013 Jul;41(Web Server issue):W89-97. doi: 10.1093/nar/gkt386. Epub 2013 May 10. Nucleic Acids Res. 2013. PMID: 23666626 Free PMC article.
-
GWENA: gene co-expression networks analysis and extended modules characterization in a single Bioconductor package.BMC Bioinformatics. 2021 May 25;22(1):267. doi: 10.1186/s12859-021-04179-4. BMC Bioinformatics. 2021. PMID: 34034647 Free PMC article.
-
RedeR: R/Bioconductor package for representing modular structures, nested networks and multiple levels of hierarchical associations.Genome Biol. 2012 Apr 24;13(4):R29. doi: 10.1186/gb-2012-13-4-r29. Genome Biol. 2012. PMID: 22531049 Free PMC article.
-
mAPKL: R/ Bioconductor package for detecting gene exemplars and revealing their characteristics.BMC Bioinformatics. 2015 Sep 15;16(1):291. doi: 10.1186/s12859-015-0719-5. BMC Bioinformatics. 2015. PMID: 26374744 Free PMC article.
Cited by
-
A data-driven network model of primary myelofibrosis: transcriptional and post-transcriptional alterations in CD34+ cells.Blood Cancer J. 2016 Jun 24;6(6):e439. doi: 10.1038/bcj.2016.47. Blood Cancer J. 2016. PMID: 27341078 Free PMC article.
-
Circulating Cell-Free Nucleic Acids as Epigenetic Biomarkers in Precision Medicine.Front Genet. 2020 Aug 11;11:844. doi: 10.3389/fgene.2020.00844. eCollection 2020. Front Genet. 2020. PMID: 32849827 Free PMC article. Review.
-
Using prior knowledge from cellular pathways and molecular networks for diagnostic specimen classification.Brief Bioinform. 2016 May;17(3):440-52. doi: 10.1093/bib/bbv044. Epub 2015 Jul 2. Brief Bioinform. 2016. PMID: 26141830 Free PMC article.
-
Systems biology, bioinformatics, and biomarkers in neuropsychiatry.Front Neurosci. 2012 Dec 24;6:187. doi: 10.3389/fnins.2012.00187. eCollection 2012. Front Neurosci. 2012. PMID: 23269912 Free PMC article.
-
Identifying cancer-related microRNAs based on subpathways.IET Syst Biol. 2018 Dec;12(6):273-278. doi: 10.1049/iet-syb.2018.5025. IET Syst Biol. 2018. PMID: 30472691 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

