In vivo screening of modified siRNAs for non-specific antiviral effect in a small fish model: number and localization in the strands are important
- PMID: 22287630
- PMCID: PMC3378874
- DOI: 10.1093/nar/gks033
In vivo screening of modified siRNAs for non-specific antiviral effect in a small fish model: number and localization in the strands are important
Abstract
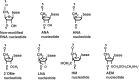
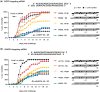
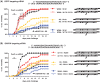
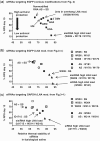
Small interfering RNAs (siRNAs) are promising new active compounds in gene medicine but the induction of non-specific immune responses following their delivery continues to be a serious problem. With the purpose of avoiding such effects chemically modified siRNAs are tested in screening assay but often only examining the expression of specific immunologically relevant genes in selected cell populations typically blood cells from treated animals or humans. Assays using a relevant physiological state in biological models as read-out are not common. Here we use a fish model where the innate antiviral effect of siRNAs is functionally monitored as reduced mortality in challenge studies involving an interferon sensitive virus. Modifications with locked nucleic acid (LNA), altritol nucleic acid (ANA) and hexitol nucleic acid (HNA) reduced the antiviral protection in this model indicative of altered immunogenicity. For LNA modified siRNAs, the number and localization of modifications in the single strands was found to be important and a correlation between antiviral protection and the thermal stability of siRNAs was found. The previously published sisiRNA will in some sequences, but not all, increase the antiviral effect of siRNAs. The applied fish model represents a potent tool for conducting fast but statistically and scientifically relevant evaluations of chemically optimized siRNAs with respect to non-specific antiviral effects in vivo.
Figures



Similar articles
-
Design of LNA-modified siRNAs against the highly structured 5' UTR of coxsackievirus B3.FEBS Lett. 2008 Sep 3;582(20):3061-6. doi: 10.1016/j.febslet.2008.07.051. Epub 2008 Aug 6. FEBS Lett. 2008. PMID: 18691577
-
LNA incorporated siRNAs exhibit lower off-target effects compared to 2'-OMethoxy in cell phenotypic assays and microarray analysis.Nucleic Acids Symp Ser (Oxf). 2008;(52):25-6. doi: 10.1093/nass/nrn013. Nucleic Acids Symp Ser (Oxf). 2008. PMID: 18776235
-
Inhibition of picornaviruses by means of RNA interference.Nucleic Acids Symp Ser (Oxf). 2008;(52):63-4. doi: 10.1093/nass/nrn032. Nucleic Acids Symp Ser (Oxf). 2008. PMID: 18776254
-
The therapeutic potential of LNA-modified siRNAs: reduction of off-target effects by chemical modification of the siRNA sequence.Methods Mol Biol. 2009;487:189-203. doi: 10.1007/978-1-60327-547-7_9. Methods Mol Biol. 2009. PMID: 19301648 Review.
-
Antiviral applications of RNAi.Curr Opin Mol Ther. 2006 Apr;8(2):115-21. Curr Opin Mol Ther. 2006. PMID: 16610763 Review.
Cited by
-
Megalin-mediated specific uptake of chitosan/siRNA nanoparticles in mouse kidney proximal tubule epithelial cells enables AQP1 gene silencing.Theranostics. 2014 Aug 13;4(10):1039-51. doi: 10.7150/thno.7866. eCollection 2014. Theranostics. 2014. PMID: 25157280 Free PMC article.
-
In vitro inhibition of Cyprinid herpesvirus-3 replication by RNAi.J Virol Methods. 2014 Sep;206(100):63-6. doi: 10.1016/j.jviromet.2014.05.022. Epub 2014 Jun 2. J Virol Methods. 2014. PMID: 24893110 Free PMC article.
-
RNA nanostructures for targeted drug delivery and imaging.RNA Biol. 2024 Jan;21(1):1-19. doi: 10.1080/15476286.2024.2328440. Epub 2024 Mar 31. RNA Biol. 2024. PMID: 38555519 Free PMC article. Review.
-
Gene therapy and kidney diseases.Mol Ther Methods Clin Dev. 2024 Sep 6;32(4):101333. doi: 10.1016/j.omtm.2024.101333. eCollection 2024 Dec 12. Mol Ther Methods Clin Dev. 2024. PMID: 39434922 Free PMC article. Review.
-
Inhibition of spring viraemia of carp virus replication in an Epithelioma papulosum cyprini cell line by RNAi.J Fish Dis. 2015 Feb;38(2):197-207. doi: 10.1111/jfd.12227. Epub 2014 Jan 27. J Fish Dis. 2015. PMID: 24460815 Free PMC article.
References
-
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. - PubMed
-
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. - PubMed
-
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. - PubMed
-
- Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA Biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. - PubMed

