NT-PGC-1α protein is sufficient to link β3-adrenergic receptor activation to transcriptional and physiological components of adaptive thermogenesis
- PMID: 22282499
- PMCID: PMC3308807
- DOI: 10.1074/jbc.M111.320200
NT-PGC-1α protein is sufficient to link β3-adrenergic receptor activation to transcriptional and physiological components of adaptive thermogenesis
Abstract
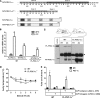
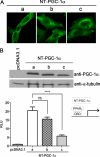
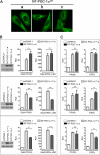
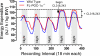
PGC-1α is an inducible transcriptional coactivator that regulates cellular energy metabolism and adaptation to environmental and nutritional stimuli. In tissues expressing PGC-1α, alternative splicing produces a truncated protein (NT-PGC-1α) corresponding to the first 267 amino acids of PGC-1α. Brown adipose tissue also expresses two novel exon 1b-derived isoforms of PGC-1α and NT-PGC-1α, which are 4 and 13 amino acids shorter in the N termini than canonical PGC-1α and NT-PGC-1α, respectively. To evaluate the ability of NT-PGC-1α to substitute for PGC-1α and assess the isoform-specific role of NT-PGC-1α, adaptive thermogenic responses of adipose tissue were evaluated in mice lacking full-length PGC-1α (FL-PGC-1(-/-)) but expressing slightly shorter but functionally equivalent forms of NT-PGC-1α (NT-PGC-1α(254)). At room temperature, NT-PGC-1α and NT-PGC-1α(254) were produced from conventional exon 1a-derived transcripts in brown adipose tissue of wild type and FL-PGC-1α(-/-) mice, respectively. However, cold exposure shifted transcription to exon 1b, increasing exon 1b-derived mRNA levels. The resulting transcriptional responses produced comparable increases in energy expenditure and maintenance of core body temperature in WT and FL-PGC-1α(-/-) mice. Moreover, treatment of the two genotypes with a selective β(3)-adrenergic receptor agonist produced similar increases in energy expenditure, mitochondrial DNA, and reductions in adiposity. Collectively, these findings illustrate that the transcriptional and physiological responses to sympathetic input are unabridged in FL-PGC-1α(-/-) mice, and that NT-PGC-1α is sufficient to link β(3)-androgenic receptor activation to adaptive thermogenesis in adipose tissue. Furthermore, the transcriptional shift from exon 1a to 1b supports isoform-specific roles for NT-PGC-1α in basal and adaptive thermogenesis.
Figures

Similar articles
-
Regulation of Brown and White Adipocyte Transcriptome by the Transcriptional Coactivator NT-PGC-1α.PLoS One. 2016 Jul 25;11(7):e0159990. doi: 10.1371/journal.pone.0159990. eCollection 2016. PLoS One. 2016. PMID: 27454177 Free PMC article.
-
NT-PGC-1α activation attenuates high-fat diet-induced obesity by enhancing brown fat thermogenesis and adipose tissue oxidative metabolism.Diabetes. 2014 Nov;63(11):3615-25. doi: 10.2337/db13-1837. Epub 2014 May 21. Diabetes. 2014. PMID: 24848065 Free PMC article.
-
NT-PGC-1α deficiency decreases mitochondrial FA oxidation in brown adipose tissue and alters substrate utilization in vivo.J Lipid Res. 2018 Sep;59(9):1660-1670. doi: 10.1194/jlr.M085647. Epub 2018 Jul 19. J Lipid Res. 2018. PMID: 30026188 Free PMC article.
-
The hitchhiker's guide to PGC-1α isoform structure and biological functions.Diabetologia. 2015 Sep;58(9):1969-77. doi: 10.1007/s00125-015-3671-z. Epub 2015 Jun 25. Diabetologia. 2015. PMID: 26109214 Review.
-
An emerging role for epigenetic regulation of Pgc-1α expression in environmentally stimulated brown adipose thermogenesis.Environ Epigenet. 2017 Jun 30;3(2):dvx009. doi: 10.1093/eep/dvx009. eCollection 2017 May. Environ Epigenet. 2017. PMID: 29492311 Free PMC article. Review.
Cited by
-
Peroxisome Proliferator-activated Receptor γ Coactivator-1 α Isoforms Selectively Regulate Multiple Splicing Events on Target Genes.J Biol Chem. 2016 Jul 15;291(29):15169-84. doi: 10.1074/jbc.M115.705822. Epub 2016 May 26. J Biol Chem. 2016. PMID: 27231350 Free PMC article.
-
Epigenetic Modifications of the PGC-1α Promoter during Exercise Induced Expression in Mice.PLoS One. 2015 Jun 8;10(6):e0129647. doi: 10.1371/journal.pone.0129647. eCollection 2015. PLoS One. 2015. PMID: 26053857 Free PMC article.
-
Adipose tissue plasticity from WAT to BAT and in between.Biochim Biophys Acta. 2014 Mar;1842(3):358-69. doi: 10.1016/j.bbadis.2013.05.011. Epub 2013 May 17. Biochim Biophys Acta. 2014. PMID: 23688783 Free PMC article. Review.
-
Transcriptional coactivator NT-PGC-1α promotes gluconeogenic gene expression and enhances hepatic gluconeogenesis.Physiol Rep. 2016 Oct;4(20):e13013. doi: 10.14814/phy2.13013. Physiol Rep. 2016. PMID: 27798359 Free PMC article.
-
Peroxisome proliferator-activated receptor gamma coactivator-1 (PGC-1) family in physiological and pathophysiological process and diseases.Signal Transduct Target Ther. 2024 Mar 1;9(1):50. doi: 10.1038/s41392-024-01756-w. Signal Transduct Target Ther. 2024. PMID: 38424050 Free PMC article. Review.
References
-
- Puigserver P., Wu Z., Park C. W., Graves R., Wright M., Spiegelman B. M. (1998) A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92, 829–839 - PubMed
-
- Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R. C., Spiegelman B. M. (1999) Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98, 115–124 - PubMed
-
- Mootha V. K., Lindgren C. M., Eriksson K. F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., Houstis N., Daly M. J., Patterson N., Mesirov J. P., Golub T. R., Tamayo P., Spiegelman B., Lander E. S., Hirschhorn J. N., Altshuler D., Groop L. C. (2003) PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately down-regulated in human diabetes. Nat. Genet. 34, 267–273 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

