SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication
- PMID: 22279047
- PMCID: PMC3273839
- DOI: 10.1101/gad.178459.111
SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication
Abstract
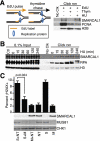
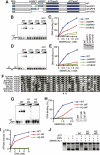
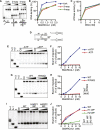
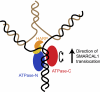
SMARCAL1 (SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily A-like1) maintains genome integrity during DNA replication. Here we investigated its mechanism of action. We found that SMARCAL1 travels with elongating replication forks, and its absence leads to MUS81-dependent double-strand break formation. Binding to specific nucleic acid substrates activates SMARCAL1 activity in a reaction that requires its HARP2 (Hep-A-related protein 2) domain. Homology modeling indicates that the HARP domain is similar in structure to the DNA-binding domain of the PUR proteins. Limited proteolysis, small-angle X-ray scattering, and functional assays indicate that the core enzymatic unit consists of the HARP2 and ATPase domains that fold into a stable structure. Surprisingly, SMARCAL1 is capable of binding three-way and four-way Holliday junctions and model replication forks that lack a designed ssDNA region. Furthermore, SMARCAL1 remodels these DNA substrates by promoting branch migration and fork regression. SMARCAL1 mutations that cause Schimke immunoosseous dysplasia or that inactivate the HARP2 domain abrogate these activities. These results suggest that SMARCAL1 continuously surveys replication forks for damage. If damage is present, it remodels the fork to promote repair and restart. Failures in the process lead to activation of an alternative repair mechanism that depends on MUS81-catalyzed cleavage of the damaged fork.
Figures




Similar articles
-
Identification and characterization of SMARCAL1 protein complexes.PLoS One. 2013 May 9;8(5):e63149. doi: 10.1371/journal.pone.0063149. Print 2013. PLoS One. 2013. PMID: 23671665 Free PMC article.
-
The HIRAN domain of helicase-like transcription factor positions the DNA translocase motor to drive efficient DNA fork regression.J Biol Chem. 2018 Jun 1;293(22):8484-8494. doi: 10.1074/jbc.RA118.002905. Epub 2018 Apr 11. J Biol Chem. 2018. PMID: 29643183 Free PMC article.
-
High-affinity DNA-binding domains of replication protein A (RPA) direct SMARCAL1-dependent replication fork remodeling.J Biol Chem. 2015 Feb 13;290(7):4110-7. doi: 10.1074/jbc.M114.627083. Epub 2014 Dec 31. J Biol Chem. 2015. PMID: 25552480 Free PMC article.
-
[SMARCAL1, roles and mechanisms in genome stability maintenance].Yi Chuan. 2019 Dec 20;41(12):1084-1098. doi: 10.16288/j.yczz.19-158. Yi Chuan. 2019. PMID: 31857280 Review. Chinese.
-
The role of SMARCAL1 in replication fork stability and telomere maintenance.DNA Repair (Amst). 2017 Aug;56:129-134. doi: 10.1016/j.dnarep.2017.06.015. Epub 2017 Jun 10. DNA Repair (Amst). 2017. PMID: 28623093 Review.
Cited by
-
Structure of a Novel DNA-binding Domain of Helicase-like Transcription Factor (HLTF) and Its Functional Implication in DNA Damage Tolerance.J Biol Chem. 2015 May 22;290(21):13215-23. doi: 10.1074/jbc.M115.643643. Epub 2015 Apr 9. J Biol Chem. 2015. PMID: 25858588 Free PMC article.
-
The cancer therapeutic potential of Chk1 inhibitors: how mechanistic studies impact on clinical trial design.Br J Clin Pharmacol. 2013 Sep;76(3):358-69. doi: 10.1111/bcp.12139. Br J Clin Pharmacol. 2013. PMID: 23593991 Free PMC article. Review.
-
Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes.Cell. 2013 Aug 1;154(3):490-503. doi: 10.1016/j.cell.2013.07.011. Cell. 2013. PMID: 23911317 Free PMC article. Review.
-
WITHDRAWN: Strand dependent bypass of DNA lesions during fork reversal by ATP-dependent translocases SMARCAL1, ZRANB3, and HLTF.bioRxiv [Preprint]. 2024 Dec 27:2024.09.17.613558. doi: 10.1101/2024.09.17.613558. bioRxiv. 2024. PMID: 39345618 Free PMC article. Preprint.
-
Role of Cockayne Syndrome Group B Protein in Replication Stress: Implications for Cancer Therapy.Int J Mol Sci. 2022 Sep 6;23(18):10212. doi: 10.3390/ijms231810212. Int J Mol Sci. 2022. PMID: 36142121 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
