Diverse functions of distal regulatory elements at the IFNG locus
- PMID: 22246629
- PMCID: PMC3273639
- DOI: 10.4049/jimmunol.1102879
Diverse functions of distal regulatory elements at the IFNG locus
Abstract
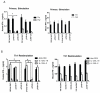
Previous studies have identified multiple conserved noncoding sequences (CNS) at the mouse Ifng locus sufficient for enhancer activity in cell-based assays. These studies do not directly address biology of the human IFNG locus in a genomic setting. IFNG enhancers may be functionally redundant or each may be functionally unique. We test the hypothesis that each IFNG enhancer has a unique necessary function using a bacterial artificial chromosome transgenic model. We find that CNS-30, CNS-4, and CNS+20 are required at distinct stages of Th1 differentiation, whereas CNS-16 has a repressive role in Th1 and Th2 cells. CNS+20 is required for IFN-γ expression by memory Th1 cells and NKT cells. CNS-4 is required for IFN-γ expression by effector Th1 cells. In contrast, CNS-16, CNS-4, and CNS+20 are each partially required for human IFN-γ expression by NK cells. Thus, IFNG CNS enhancers have redundant necessary functions in NK cells but unique necessary functions in Th cells. These results also demonstrate that distinct CNSs are required to transcribe IFNG at each stage of the Th1 differentiation pathway.
Figures

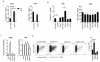
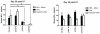
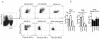

Similar articles
-
Distal regions of the human IFNG locus direct cell type-specific expression.J Immunol. 2010 Aug 1;185(3):1492-501. doi: 10.4049/jimmunol.1000124. Epub 2010 Jun 23. J Immunol. 2010. PMID: 20574006 Free PMC article.
-
Loss of methylation at the IFNG promoter and CNS-1 is associated with the development of functional IFN-γ memory in human CD4(+) T lymphocytes.Eur J Immunol. 2013 Mar;43(3):793-804. doi: 10.1002/eji.201242858. Eur J Immunol. 2013. PMID: 23255246
-
Deletion of a conserved cis-element in the Ifng locus highlights the role of acute histone acetylation in modulating inducible gene transcription.PLoS Genet. 2014 Jan;10(1):e1003969. doi: 10.1371/journal.pgen.1003969. Epub 2014 Jan 9. PLoS Genet. 2014. PMID: 24415943 Free PMC article.
-
BACing up the interferon-gamma locus.Immunity. 2006 Nov;25(5):691-3. doi: 10.1016/j.immuni.2006.10.006. Immunity. 2006. PMID: 17098198 Free PMC article. Review.
-
Regulation of interferon-gamma during innate and adaptive immune responses.Adv Immunol. 2007;96:41-101. doi: 10.1016/S0065-2776(07)96002-2. Adv Immunol. 2007. PMID: 17981204 Review.
Cited by
-
Epigenetic Activation and Silencing of the Gene that Encodes IFN-γ.Front Immunol. 2013 May 16;4:112. doi: 10.3389/fimmu.2013.00112. eCollection 2013. Front Immunol. 2013. PMID: 23720660 Free PMC article.
-
Human invariant natural killer T cells acquire transient innate responsiveness via histone H4 acetylation induced by weak TCR stimulation.J Exp Med. 2012 May 7;209(5):987-1000. doi: 10.1084/jem.20111024. Epub 2012 Apr 16. J Exp Med. 2012. PMID: 22508835 Free PMC article.
-
Long noncoding RNAs in T lymphocytes.J Leukoc Biol. 2016 Jan;99(1):31-44. doi: 10.1189/jlb.1RI0815-389R. Epub 2015 Nov 4. J Leukoc Biol. 2016. PMID: 26538526 Free PMC article. Review.
-
Distinct Gene Regulatory Pathways for Human Innate versus Adaptive Lymphoid Cells.Cell. 2016 May 19;165(5):1134-1146. doi: 10.1016/j.cell.2016.04.014. Epub 2016 May 5. Cell. 2016. PMID: 27156452 Free PMC article.
-
Long non-coding RNAs in innate and adaptive immunity.Virus Res. 2016 Jan 2;212:146-60. doi: 10.1016/j.virusres.2015.07.003. Epub 2015 Jul 9. Virus Res. 2016. PMID: 26166759 Free PMC article. Review.
References
-
- Murphy KM, Reiner SL. The lineage decisions of helper T cells. Nat Rev Immunol. 2002;2:933–944. - PubMed
-
- Ansel KM, Lee DU, Rao A. An epigenetic view of helper T cell differentiation. Nat Immunol. 2003;4:616–623. - PubMed
-
- Soutto M, Zhou W, Aune TM. Cutting edge: distal regulatory elements are required to achieve selective expression of IFN-gamma in Th1/Tc1 effector cells. J Immunol. 2002;169:6664–6667. - PubMed
-
- Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363:166–176. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI044924-09A1/AI/NIAID NIH HHS/United States
- P30 DK058404/DK/NIDDK NIH HHS/United States
- TL1 RR024978/RR/NCRR NIH HHS/United States
- KL2 RR024977/RR/NCRR NIH HHS/United States
- R01 AI044924-10/AI/NIAID NIH HHS/United States
- T32 HL069765-07/HL/NHLBI NIH HHS/United States
- T32 HL069765/HL/NHLBI NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- HL069765/HL/NHLBI NIH HHS/United States
- DK058404/DK/NIDDK NIH HHS/United States
- 5UL1 RR024975-03/RR/NCRR NIH HHS/United States
- R56 AI044924/AI/NIAID NIH HHS/United States
- CA68485/CA/NCI NIH HHS/United States
- UL1 RR024975/RR/NCRR NIH HHS/United States
- AI44924/AI/NIAID NIH HHS/United States
- R01 AI044924/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

