Clinical significance of miR-155 expression in breast cancer and effects of miR-155 ASO on cell viability and apoptosis
- PMID: 22245916
- PMCID: PMC3583512
- DOI: 10.3892/or.2012.1634
Clinical significance of miR-155 expression in breast cancer and effects of miR-155 ASO on cell viability and apoptosis
Abstract
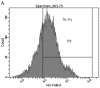
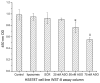
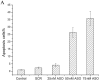
Accumulating evidence shows that mircroRNAs (miRNAs) play a vital role in tumorigenesis. miR-155 is one of the most multifunctional miRNAs whose overexpression has been found to be associated with different types of cancer including breast cancer. To further determine the potential involvement of miR-155 in breast cancer, we evaluated the expression levels of miR-155 by real-time PCR and correlated the results with clinicopathological features. Matched non-tumor and tumor tissues of 42 infiltrating ductal carcinomas and 3 infiltrating lobular carcinomas were analyzed for miR-155 expression by real-time PCR. Further, we used an antisense technique to inhibit miR-155 expression in vitro. WST-8 test was performed to evaluate cell viability and apoptosis assay was used to investigate the effect of the miR-155 antisense oligonucleotide (miR-155 ASO) on HS578T cell death. The expression levels of miR-155 were significantly higher in tumor tissues than the levels in matched non-tumor tissues (P<0.001). Up-regulated miR-155 expression was associated with lymph node positivity (P=0.034), higher proliferation index (Ki-67 >10%) (P=0.019) and advanced breast cancer TNM clinical stage (P=0.002). Interestingly, we next found that miR-155 expression levels had close relations with ER status (P=0.041) and PR status (P=0.029). Transfection efficiency detected by flow cytometry was higher than 70%, the WST-8 test showed that viability of HS578T cells was greatly reduced after transfection with miR-155 ASO compared with the scramble (SCR) group or the liposome group. The Annexin V-FITC/PI assay also indicated that transfection with miR-155 ASO promoted apoptosis.
Figures









Similar articles
-
Mitochondrial UCP4 and bcl-2 expression in imprints of breast carcinomas: relationship with DNA ploidy and classical prognostic factors.Pathol Res Pract. 2011 Jun 15;207(6):377-82. doi: 10.1016/j.prp.2011.03.007. Epub 2011 May 31. Pathol Res Pract. 2011. PMID: 21621926
-
The difference in miR-21 expression levels between invasive and non-invasive breast cancers emphasizes its role in breast cancer invasion.Med Oncol. 2014 Mar;31(3):867. doi: 10.1007/s12032-014-0867-x. Epub 2014 Feb 2. Med Oncol. 2014. PMID: 24488617
-
Tumor-suppressive microRNA-34a inhibits breast cancer cell migration and invasion via targeting oncogenic TPD52.Tumour Biol. 2016 Jun;37(6):7481-91. doi: 10.1007/s13277-015-4623-4. Epub 2015 Dec 17. Tumour Biol. 2016. PMID: 26678891
-
Prognostic and biological significance of microRNA-127 expression in human breast cancer.Dis Markers. 2014;2014:401986. doi: 10.1155/2014/401986. Epub 2014 Nov 12. Dis Markers. 2014. PMID: 25477702 Free PMC article.
-
Higher miR-21 expression in invasive breast carcinomas is associated with positive estrogen and progesterone receptor status in patients from Serbia.Med Oncol. 2014 Jun;31(6):977. doi: 10.1007/s12032-014-0977-5. Epub 2014 Apr 30. Med Oncol. 2014. PMID: 24781337
Cited by
-
MicroRNA-155 complementation on a chemically functionalized dual electrode surface for determining breast cancer.3 Biotech. 2020 Jun;10(6):270. doi: 10.1007/s13205-020-02261-x. Epub 2020 May 28. 3 Biotech. 2020. PMID: 32523864 Free PMC article.
-
Non-Coding RNAs Regulate Spinal Cord Injury-Related Neuropathic Pain via Neuroinflammation.J Inflamm Res. 2023 Jun 13;16:2477-2489. doi: 10.2147/JIR.S413264. eCollection 2023. J Inflamm Res. 2023. PMID: 37334347 Free PMC article. Review.
-
Unraveling the Potential of miRNAs from CSCs as an Emerging Clinical Tool for Breast Cancer Diagnosis and Prognosis.Int J Mol Sci. 2023 Nov 6;24(21):16010. doi: 10.3390/ijms242116010. Int J Mol Sci. 2023. PMID: 37958993 Free PMC article. Review.
-
High expression of miR-21 in triple-negative breast cancers was correlated with a poor prognosis and promoted tumor cell in vitro proliferation.Med Oncol. 2014 Jul;31(7):57. doi: 10.1007/s12032-014-0057-x. Epub 2014 Jun 15. Med Oncol. 2014. PMID: 24930006
-
Clinical Evaluation of the Diagnostic Role of MicroRNA-155 in Breast Cancer.Int J Genomics. 2020 Sep 7;2020:9514831. doi: 10.1155/2020/9514831. eCollection 2020. Int J Genomics. 2020. PMID: 32964011 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

