Bacterial iron-sulfur regulatory proteins as biological sensor-switches
- PMID: 22239203
- PMCID: PMC3430481
- DOI: 10.1089/ars.2012.4511
Bacterial iron-sulfur regulatory proteins as biological sensor-switches
Abstract
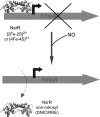
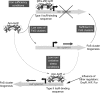
Significance: In recent years, bacterial iron-sulfur cluster proteins that function as regulators of gene transcription have emerged as a major new group. In all cases, the cluster acts as a sensor of the environment and enables the organism to adapt to the prevailing conditions. This can range from mounting a response to oxidative or nitrosative stress to switching between anaerobic and aerobic respiratory pathways. The sensitivity of these ancient cofactors to small molecule reactive oxygen and nitrogen species, in particular, makes them ideally suited to function as sensors.
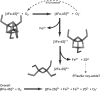
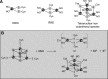
Recent advances: An important challenge is to obtain mechanistic and structural information about how these regulators function and, in particular, how the chemistry occurring at the cluster drives the subsequent regulatory response. For several regulators, including FNR, SoxR, NsrR, IscR, and Wbl proteins, major advances in understanding have been gained recently and these are reviewed here.
Critical issues: A common theme emerging from these studies is that the sensitivity and specificity of the cluster of each regulatory protein must be exquisitely controlled by the protein environment of the cluster.
Future directions: A major future challenge is to determine, for a range of regulators, the key factors for achieving control of sensitivity/specificity. Such information will lead, eventually, to a system understanding of stress response, which often involves more than one regulator.
Figures
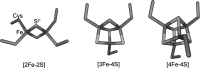



Similar articles
-
Bacterial iron-sulfur cluster sensors in mammalian pathogens.Metallomics. 2015 Jun;7(6):943-56. doi: 10.1039/c5mt00012b. Metallomics. 2015. PMID: 25738802 Free PMC article. Review.
-
Oxidative stress sensing by the iron-sulfur cluster in the transcription factor, SoxR.J Inorg Biochem. 2014 Apr;133:87-91. doi: 10.1016/j.jinorgbio.2013.11.008. Epub 2013 Nov 28. J Inorg Biochem. 2014. PMID: 24332474 Review.
-
Redox-Sensing Iron-Sulfur Cluster Regulators.Antioxid Redox Signal. 2018 Dec 20;29(18):1809-1829. doi: 10.1089/ars.2017.7361. Epub 2017 Nov 7. Antioxid Redox Signal. 2018. PMID: 28967283 Review.
-
Redox-sensing iron-sulfur cluster regulators.Antioxid Redox Signal. 2017 Dec 7. doi: 10.1089/ars.2017.7369. Online ahead of print. Antioxid Redox Signal. 2017. PMID: 29216742
-
A Regulatory Circuit Composed of a Transcription Factor, IscR, and a Regulatory RNA, RyhB, Controls Fe-S Cluster Delivery.mBio. 2016 Sep 20;7(5):e00966-16. doi: 10.1128/mBio.00966-16. mBio. 2016. PMID: 27651365 Free PMC article.
Cited by
-
FeoC from Klebsiella pneumoniae contains a [4Fe-4S] cluster.J Bacteriol. 2013 Oct;195(20):4726-34. doi: 10.1128/JB.00687-13. Epub 2013 Aug 16. J Bacteriol. 2013. PMID: 23955005 Free PMC article.
-
Global transcriptional regulator FNR regulates the pyruvate cycle and proton motive force to play a role in aminoglycosides resistance of Edwardsiella tarda.Front Microbiol. 2022 Sep 7;13:1003586. doi: 10.3389/fmicb.2022.1003586. eCollection 2022. Front Microbiol. 2022. PMID: 36160231 Free PMC article.
-
A Review: Origins of the Dielectric Properties of Proteins and Potential Development as Bio-Sensors.Sensors (Basel). 2016 Aug 4;16(8):1232. doi: 10.3390/s16081232. Sensors (Basel). 2016. PMID: 27527179 Free PMC article. Review.
-
Fe-S proteins that regulate gene expression.Biochim Biophys Acta. 2015 Jun;1853(6):1284-93. doi: 10.1016/j.bbamcr.2014.11.018. Epub 2014 Nov 20. Biochim Biophys Acta. 2015. PMID: 25450978 Free PMC article. Review.
-
The basic leucine zipper stress response regulator Yap5 senses high-iron conditions by coordination of [2Fe-2S] clusters.Mol Cell Biol. 2015 Jan;35(2):370-8. doi: 10.1128/MCB.01033-14. Epub 2014 Nov 3. Mol Cell Biol. 2015. PMID: 25368382 Free PMC article.
References
-
- Achebach S. Selmer T. Unden G. Properties and significance of apo FNR as a second form of air-inactivated [4Fe-4S] FNR of Escherichia coli. FEBS J. 2005;272:4260–4269. - PubMed
-
- Alam MS. Garg SK. Agrawal P. Molecular function of WhiB4/Rv3681c of Mycobacterium tuberculosis H37Rv: a [4Fe-4S] cluster co-ordinating protein disulphide reductase. Mol Microbiol. 2007;63:1414–1431. - PubMed
-
- Arnon DI. Whatley FR. Allen MB. Triphosphopyridine nucleotide as a catalyst of photosynthetic phosphorylation. Nature. 1957;180:182–185. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/G018960/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 078731/Z/05/Z/WT_/Wellcome Trust/United Kingdom
- BB/D00926X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/G019347/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/J/00000015/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources

