Connecting the dots between PubMed abstracts
- PMID: 22235301
- PMCID: PMC3250456
- DOI: 10.1371/journal.pone.0029509
Connecting the dots between PubMed abstracts
Abstract
Background: There are now a multitude of articles published in a diversity of journals providing information about genes, proteins, pathways, and diseases. Each article investigates subsets of a biological process, but to gain insight into the functioning of a system as a whole, we must integrate information from multiple publications. Particularly, unraveling relationships between extra-cellular inputs and downstream molecular response mechanisms requires integrating conclusions from diverse publications.
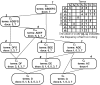
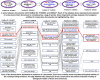
Methodology: We present an automated approach to biological knowledge discovery from PubMed abstracts, suitable for "connecting the dots" across the literature. We describe a storytelling algorithm that, given a start and end publication, typically with little or no overlap in content, identifies a chain of intermediate publications from one to the other, such that neighboring publications have significant content similarity. The quality of discovered stories is measured using local criteria such as the size of supporting neighborhoods for each link and the strength of individual links connecting publications, as well as global metrics of dispersion. To ensure that the story stays coherent as it meanders from one publication to another, we demonstrate the design of novel coherence and overlap filters for use as post-processing steps.
Conclusions: WE DEMONSTRATE THE APPLICATION OF OUR STORYTELLING ALGORITHM TO THREE CASE STUDIES: i) a many-one study exploring relationships between multiple cellular inputs and a molecule responsible for cell-fate decisions, ii) a many-many study exploring the relationships between multiple cytokines and multiple downstream transcription factors, and iii) a one-to-one study to showcase the ability to recover a cancer related association, viz. the Warburg effect, from past literature. The storytelling pipeline helps narrow down a scientist's focus from several hundreds of thousands of relevant documents to only around a hundred stories. We argue that our approach can serve as a valuable discovery aid for hypothesis generation and connection exploration in large unstructured biological knowledge bases.
Conflict of interest statement
Figures


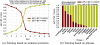



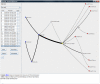
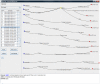
 , and (b) shows the dispersion plot of a story with dispersion coefficient
, and (b) shows the dispersion plot of a story with dispersion coefficient  .
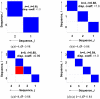
.



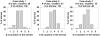
Similar articles
-
pubmed2ensembl: a resource for mining the biological literature on genes.PLoS One. 2011;6(9):e24716. doi: 10.1371/journal.pone.0024716. Epub 2011 Sep 29. PLoS One. 2011. PMID: 21980353 Free PMC article.
-
OmixLitMiner: A Bioinformatics Tool for Prioritizing Biological Leads from 'Omics Data Using Literature Retrieval and Data Mining.Int J Mol Sci. 2020 Feb 19;21(4):1374. doi: 10.3390/ijms21041374. Int J Mol Sci. 2020. PMID: 32092871 Free PMC article.
-
BioTextQuest(+): a knowledge integration platform for literature mining and concept discovery.Bioinformatics. 2014 Nov 15;30(22):3249-56. doi: 10.1093/bioinformatics/btu524. Epub 2014 Aug 6. Bioinformatics. 2014. PMID: 25100685
-
HEALTH GeoJunction: place-time-concept browsing of health publications.Int J Health Geogr. 2010 May 18;9:23. doi: 10.1186/1476-072X-9-23. Int J Health Geogr. 2010. PMID: 20482806 Free PMC article. Review.
-
Translational Metabolomics of Head Injury: Exploring Dysfunctional Cerebral Metabolism with Ex Vivo NMR Spectroscopy-Based Metabolite Quantification.In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. PMID: 26269925 Free Books & Documents. Review.
Cited by
-
Narratives in the network: interactive methods for mining cell signaling networks.J Comput Biol. 2012 Sep;19(9):1043-59. doi: 10.1089/cmb.2011.0244. Epub 2012 Aug 16. J Comput Biol. 2012. PMID: 22897227 Free PMC article.
-
A systematic review on literature-based discovery workflow.PeerJ Comput Sci. 2019 Nov 18;5:e235. doi: 10.7717/peerj-cs.235. eCollection 2019. PeerJ Comput Sci. 2019. PMID: 33816888 Free PMC article.
-
Rediscovering Don Swanson: the Past, Present and Future of Literature-Based Discovery.J Data Inf Sci. 2017 Dec;2(4):43-64. doi: 10.1515/jdis-2017-0019. J Data Inf Sci. 2017. PMID: 29355246 Free PMC article.
References
-
- Carpenter AE, Sabatini DM. Systematic Genome-wide Screens of Gene Function. Nat Rev Genet. 2004;5:11–22. - PubMed
-
- Shatkay H, Feldman R. Mining the Biomedical Literature in the Genomic Era: an Overview. J Comput Biol. 2003;10:821–855. - PubMed
-
- Zhou D, He Y. Extracting Interactions between Proteins from the Literature. J Biomed Inform. 2008;41:393–407. - PubMed
-
- Kersey P, Apweiler R. Linking Publication, Gene and Protein Data. Nat Cell Biol. 2006;8:1183–1189. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

