DNA condensation by TmHU studied by optical tweezers, AFM and molecular dynamics simulations
- PMID: 22210966
- PMCID: PMC3006463
- DOI: 10.1007/s10867-010-9203-7
DNA condensation by TmHU studied by optical tweezers, AFM and molecular dynamics simulations
Abstract
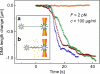
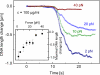
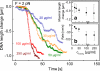
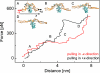
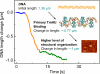
The compaction of DNA by the HU protein from Thermotoga maritima (TmHU) is analysed on a single-molecule level by the usage of an optical tweezers-assisted force clamp. The condensation reaction is investigated at forces between 2 and 40 pN applied to the ends of the DNA as well as in dependence on the TmHU concentration. At 2 and 5 pN, the DNA compaction down to 30% of the initial end-to-end distance takes place in two regimes. Increasing the force changes the progression of the reaction until almost nothing is observed at 40 pN. Based on the results of steered molecular dynamics simulations, the first regime of the length reduction is assigned to a primary level of DNA compaction by TmHU. The second one is supposed to correspond to the formation of higher levels of structural organisation. These findings are supported by results obtained by atomic force microscopy.
Keywords: Force clamp; Optical tweezers; Protein–DNA interaction; Single-molecule study; Steered molecular dynamics; Thermotoga maritima.
Figures

Similar articles
-
Kinetics of TmHU binding to DNA as observed by optical tweezers.Microsc Res Tech. 2007 Nov;70(11):938-43. doi: 10.1002/jemt.20498. Microsc Res Tech. 2007. PMID: 17661366
-
Binding of TmHU to single dsDNA as observed by optical tweezers.J Mol Biol. 2006 Jun 9;359(3):769-76. doi: 10.1016/j.jmb.2006.04.006. Epub 2006 Apr 19. J Mol Biol. 2006. PMID: 16647714
-
High-affinity DNA binding of HU protein from the hyperthermophile Thermotoga maritima.J Mol Biol. 2001 Aug 17;311(3):491-502. doi: 10.1006/jmbi.2001.4763. J Mol Biol. 2001. PMID: 11493003
-
Introduction to optical tweezers: background, system designs, and commercial solutions.Methods Mol Biol. 2011;783:1-20. doi: 10.1007/978-1-61779-282-3_1. Methods Mol Biol. 2011. PMID: 21909880 Review.
-
Single-Molecule Analysis and Engineering of DNA Motors.Chem Rev. 2020 Jan 8;120(1):36-78. doi: 10.1021/acs.chemrev.9b00361. Epub 2019 Oct 29. Chem Rev. 2020. PMID: 31661246 Review.
Cited by
-
Single-molecule imaging of genome maintenance proteins encountering specific DNA sequences and structures.DNA Repair (Amst). 2023 Aug;128:103528. doi: 10.1016/j.dnarep.2023.103528. Epub 2023 Jun 24. DNA Repair (Amst). 2023. PMID: 37392578 Free PMC article. Review.
-
ParB spreading requires DNA bridging.Genes Dev. 2014 Jun 1;28(11):1228-38. doi: 10.1101/gad.242206.114. Epub 2014 May 14. Genes Dev. 2014. PMID: 24829297 Free PMC article.
References
-
- Nelson KE, Clayton RA, Gill SR, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Nelson WC, Ketchum KA, McDonald L, Utterback TR, Malek JA, Linher KD, Garrett MM, Stewart AM, Cotton MD, Pratt MS, Phillips CA, Richardson D, Heidelberg J, Sutton GG, Fleischmann RD, Eisen JA, White O, Salzberg SL, Smith HO, Venter JC, Fraser CM.Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima Nature 1999399323–329.10.1038/206011999Natur.399..323N - DOI - PubMed
-
- Huber R, Langworthy TA, König H, Thomm M, Woese CR, Sleytr W, Stetter KO. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90°. Arch. Microbiol. 1986;144:324–333. doi: 10.1007/BF00409880. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

