Thermodynamics of ligand binding to a heterogeneous RNA population in the malachite green aptamer
- PMID: 22192051
- PMCID: PMC3267630
- DOI: 10.1021/bi201642p
Thermodynamics of ligand binding to a heterogeneous RNA population in the malachite green aptamer
Abstract
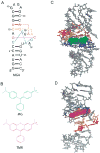
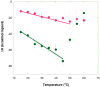
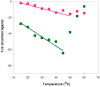
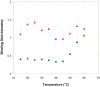
The malachite green aptamer binds two closely related ligands, malachite green (MG) and tetramethylrosamine (TMR), with nearly equal affinity. The MG ligand consists of three phenyl rings emanating from a central carbon, while TMR has two of the three rings connected by an ether linkage. The binding pockets for MG and TMR in the aptamer, known from high-resolution structures, differ only in the conformation of a few nucleotides. Herein, we applied isothermal titration calorimetry (ITC) to compare the thermodynamics of binding of MG and TMR to the aptamer. Binding heat capacities were obtained from ITC titrations over the temperature range of 15-60 °C. Two temperature regimes were found for MG binding: one from 15 to 45 °C where MG bound with a large negative heat capacity and an apparent stoichiometry (n) of ~0.4 and another from 50 to 60 °C where MG bound with a positive heat capacity and an n of ~1.1. The binding of TMR, on the other hand, revealed only one temperature regime for binding, with a more modest negative heat capacity and an n of ~1.2. The large difference in heat capacity between the two ligands suggests that significantly more conformational rearrangement occurs upon the binding of MG than that of TMR, which is consistent with differences in solvent accessible surface area calculated for available ligand-bound structures. Lastly, we note that the binding stoichiometry of MG was improved not only by an increase in the temperature but also by a decrease in the concentration of Mg(2+) or an increase in the time between ITC injections. These studies suggest that binding of a dynamical ligand to a functional RNA requires the RNA itself to have significant dynamics.
Figures

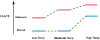
Similar articles
-
Thermodynamics and kinetics of adaptive binding in the malachite green RNA aptamer.Biochemistry. 2013 Sep 24;52(38):6575-83. doi: 10.1021/bi400549s. Epub 2013 Sep 11. Biochemistry. 2013. PMID: 23984874
-
Entropy and Mg2+ control ligand affinity and specificity in the malachite green binding RNA aptamer.Mol Biosyst. 2011 Jul;7(7):2156-63. doi: 10.1039/c1mb05075c. Epub 2011 Apr 26. Mol Biosyst. 2011. PMID: 21523267
-
2.8 A crystal structure of the malachite green aptamer.J Mol Biol. 2000 Aug 4;301(1):117-28. doi: 10.1006/jmbi.2000.3951. J Mol Biol. 2000. PMID: 10926496
-
Investigations on the interface of nucleic acid aptamers and binding targets.Analyst. 2018 Nov 5;143(22):5317-5338. doi: 10.1039/c8an01467a. Analyst. 2018. PMID: 30357118 Review.
-
Applications of isothermal titration calorimetry in RNA biochemistry and biophysics.Biopolymers. 2007 Dec 5-15;87(5-6):293-301. doi: 10.1002/bip.20816. Biopolymers. 2007. PMID: 17671974 Free PMC article. Review.
Cited by
-
Fluorescence-Based Strategies to Investigate the Structure and Dynamics of Aptamer-Ligand Complexes.Front Chem. 2016 Aug 3;4:33. doi: 10.3389/fchem.2016.00033. eCollection 2016. Front Chem. 2016. PMID: 27536656 Free PMC article. Review.
-
ITC analysis of ligand binding to preQ₁ riboswitches.Methods Enzymol. 2014;549:435-50. doi: 10.1016/B978-0-12-801122-5.00018-0. Methods Enzymol. 2014. PMID: 25432759 Free PMC article.
-
Expanding the DNA-encoded library toolbox: identifying small molecules targeting RNA.Nucleic Acids Res. 2022 Jul 8;50(12):e67. doi: 10.1093/nar/gkac173. Nucleic Acids Res. 2022. PMID: 35288754 Free PMC article.
-
Kinetic and equilibrium binding characterization of aptamers to small molecules using a label-free, sensitive, and scalable platform.Anal Chem. 2014 Apr 1;86(7):3273-8. doi: 10.1021/ac5001527. Epub 2014 Mar 14. Anal Chem. 2014. PMID: 24548121 Free PMC article.
-
Molecular crowders and cosolutes promote folding cooperativity of RNA under physiological ionic conditions.RNA. 2014 Mar;20(3):331-47. doi: 10.1261/rna.042747.113. Epub 2014 Jan 17. RNA. 2014. PMID: 24442612 Free PMC article.
References
-
- Ellington AD. RNA selection. Aptamers achieve the desired recognition. Curr Biol. 1994;4:427–429. - PubMed
-
- Joyce GF. In vitro evolution of nucleic acids. Curr Opin Struct Biol. 1994;4:331–336. - PubMed
-
- Mayer G. The chemical biology of aptamers. Angew Chem Int Ed Engl. 2009;48:2672–2689. - PubMed
-
- Giovannoli C, Baggiani C, Anfossi L, Giraudi G. Aptamers and molecularly imprinted polymers as artificial biomimetic receptors in affinity capillary electrophoresis and electrochromatography. Electrophoresis. 2008;29:3349–3365. - PubMed
-
- Cho EJ, Lee JW, Ellington AD. Applications of aptamers as sensors. Annu Rev Anal Chem. 2009;2:241–264. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

