Matrix proteases in mitochondrial DNA function
- PMID: 22172992
- PMCID: PMC3383373
- DOI: 10.1016/j.bbagrm.2011.11.008
Matrix proteases in mitochondrial DNA function
Abstract
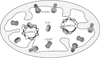
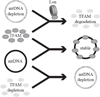
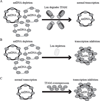
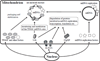
Lon, ClpXP and m-AAA are the three major ATP-dependent proteases in the mitochondrial matrix. All three are involved in general quality control by degrading damaged or abnormal proteins. In addition to this role, they are proposed to serve roles in mitochondrial DNA functions including packaging and stability, replication, transcription and translation. In particular, Lon has been implicated in mtDNA metabolism in yeast, fly and humans. Here, we review the role of Lon protease in mitochondrial DNA functions, and discuss a putative physiological role for mitochondrial transcription factor A (TFAM) degradation by Lon protease. We also discuss the possible roles of m-AAA and ClpXP in mitochondrial DNA functions, and the putative candidate substrates for the three matrix proteases. This article is part of a Special Issue entitled: Mitochondrial Gene Expression.
Copyright © 2011 Elsevier B.V. All rights reserved.
Figures

Similar articles
-
Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ Lon protease.Mol Cell. 2013 Jan 10;49(1):121-32. doi: 10.1016/j.molcel.2012.10.023. Epub 2012 Nov 29. Mol Cell. 2013. PMID: 23201127 Free PMC article.
-
The influence of ATP-dependent proteases on a variety of nucleoid-associated processes.J Struct Biol. 2012 Aug;179(2):181-92. doi: 10.1016/j.jsb.2012.05.018. Epub 2012 Jun 6. J Struct Biol. 2012. PMID: 22683345 Review.
-
Mitochondrial Lon protease regulates mitochondrial DNA copy number and transcription by selective degradation of mitochondrial transcription factor A (TFAM).Proc Natl Acad Sci U S A. 2010 Oct 26;107(43):18410-5. doi: 10.1073/pnas.1008924107. Epub 2010 Oct 7. Proc Natl Acad Sci U S A. 2010. PMID: 20930118 Free PMC article.
-
Mitochondrial transcription factor A regulates mitochondrial transcription initiation, DNA packaging, and genome copy number.Biochim Biophys Acta. 2012 Sep-Oct;1819(9-10):921-9. doi: 10.1016/j.bbagrm.2012.03.002. Epub 2012 Mar 21. Biochim Biophys Acta. 2012. PMID: 22465614 Review.
-
Maintenance of mitochondrial genome distribution by mitochondrial AAA+ protein ClpX.Exp Cell Res. 2012 Nov 1;318(18):2335-43. doi: 10.1016/j.yexcr.2012.07.012. Epub 2012 Jul 24. Exp Cell Res. 2012. PMID: 22841477
Cited by
-
The mitochondrial nucleoid: integrating mitochondrial DNA into cellular homeostasis.Cold Spring Harb Perspect Biol. 2013 May 1;5(5):a011080. doi: 10.1101/cshperspect.a011080. Cold Spring Harb Perspect Biol. 2013. PMID: 23637282 Free PMC article. Review.
-
NOA1, a novel ClpXP substrate, takes an unexpected nuclear detour prior to mitochondrial import.PLoS One. 2014 Jul 29;9(7):e103141. doi: 10.1371/journal.pone.0103141. eCollection 2014. PLoS One. 2014. PMID: 25072814 Free PMC article.
-
Mitochondrial Biogenesis in Response to Chromium (VI) Toxicity in Human Liver Cells.Int J Mol Sci. 2017 Sep 14;18(9):1877. doi: 10.3390/ijms18091877. Int J Mol Sci. 2017. PMID: 28906435 Free PMC article.
-
Energizing Genetics and Epi-genetics: Role in the Regulation of Mitochondrial Function.Curr Genomics. 2014 Dec;15(6):436-56. doi: 10.2174/138920291506150106151119. Curr Genomics. 2014. PMID: 25646072 Free PMC article.
-
A Mechanistic Review of Mitophagy and Its Role in Protection against Alcoholic Liver Disease.Biomolecules. 2015 Oct 16;5(4):2619-42. doi: 10.3390/biom5042619. Biomolecules. 2015. PMID: 26501336 Free PMC article. Review.
References
-
- Ugarte N, Petropoulos I, Friguet B. Oxidized mitochondrial protein degradation and repair in aging and oxidative stress. Antioxid Redox Signal. 2010;13:539–549. - PubMed
-
- Truscott KN, Lowth BR, Strack PR, Dougan DA. Diverse functions of mitochondrial AAA+ proteins: protein activation, disaggregation, and degradation. Biochem Cell Biol. 2010;88:97–108. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

