HSC20 interacts with frataxin and is involved in iron-sulfur cluster biogenesis and iron homeostasis
- PMID: 22171070
- PMCID: PMC3298274
- DOI: 10.1093/hmg/ddr582
HSC20 interacts with frataxin and is involved in iron-sulfur cluster biogenesis and iron homeostasis
Abstract
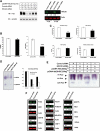
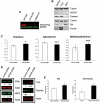
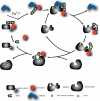
Friedreich's ataxia is a neurodegenerative disorder caused by mutations in the frataxin gene that produces a predominantly mitochondrial protein whose primary function appears to be mitochondrial iron-sulfur cluster (ISC) biosynthesis. Previously we demonstrated that frataxin interacts with multiple components of the mammalian ISC assembly machinery. Here we demonstrate that frataxin interacts with the mammalian mitochondrial chaperone HSC20. We show that this interaction is iron-dependent. We also show that like frataxin, HSC20 interacts with multiple proteins involved in ISC biogenesis including the ISCU/Nfs1 ISC biogenesis complex and the GRP75 ISC chaperone. Furthermore, knockdown of HSC20 caused functional defects in activity of mitochondrial ISC-containing enzymes and also defects in ISC protein expression. Alterations up or down of frataxin expression caused compensatory changes in HSC20 expression inversely, as expected of two cooperating proteins operating in the same pathway and suggesting a potential therapeutic strategy for the disease. Knockdown of HSC20 altered cytosolic and mitochondrial iron pools and increased the expression of transferrin receptor 1 and iron regulatory protein 2 consistent with decreased iron bioavailability. These results indicate that HSC20 interacts with frataxin structurally and functionally and is important for ISC biogenesis and iron homeostasis in mammals. Furthermore, they suggest that HSC20 may act late in the ISC pathway as a chaperone in ISC delivery to apoproteins and that HSC20 should be included in multi-protein complex studies of mammalian ISC biogenesis.
Figures

Similar articles
-
Mitochondrial frataxin interacts with ISD11 of the NFS1/ISCU complex and multiple mitochondrial chaperones.Hum Mol Genet. 2007 Apr 15;16(8):929-41. doi: 10.1093/hmg/ddm038. Epub 2007 Mar 1. Hum Mol Genet. 2007. PMID: 17331979
-
Cytosolic HSC20 integrates de novo iron-sulfur cluster biogenesis with the CIAO1-mediated transfer to recipients.Hum Mol Genet. 2018 Mar 1;27(5):837-852. doi: 10.1093/hmg/ddy004. Hum Mol Genet. 2018. PMID: 29309586 Free PMC article.
-
Insertion mutants in Drosophila melanogaster Hsc20 halt larval growth and lead to reduced iron-sulfur cluster enzyme activities and impaired iron homeostasis.J Biol Inorg Chem. 2013 Apr;18(4):441-9. doi: 10.1007/s00775-013-0988-2. Epub 2013 Feb 27. J Biol Inorg Chem. 2013. PMID: 23444034 Free PMC article.
-
Mechanisms of Mitochondrial Iron-Sulfur Protein Biogenesis.Annu Rev Biochem. 2020 Jun 20;89:471-499. doi: 10.1146/annurev-biochem-013118-111540. Epub 2020 Jan 14. Annu Rev Biochem. 2020. PMID: 31935115 Review.
-
Frataxin and mitochondrial FeS cluster biogenesis.J Biol Chem. 2010 Aug 27;285(35):26737-26743. doi: 10.1074/jbc.R110.118679. Epub 2010 Jun 3. J Biol Chem. 2010. PMID: 20522547 Free PMC article. Review.
Cited by
-
Fe-S Protein Synthesis in Green Algae Mitochondria.Plants (Basel). 2021 Jan 21;10(2):200. doi: 10.3390/plants10020200. Plants (Basel). 2021. PMID: 33494487 Free PMC article. Review.
-
Plant Frataxin in Metal Metabolism.Front Plant Sci. 2018 Nov 21;9:1706. doi: 10.3389/fpls.2018.01706. eCollection 2018. Front Plant Sci. 2018. PMID: 30519254 Free PMC article.
-
The interactions of molecular chaperones with client proteins: why are they so weak?J Biol Chem. 2021 Nov;297(5):101282. doi: 10.1016/j.jbc.2021.101282. Epub 2021 Oct 6. J Biol Chem. 2021. PMID: 34624315 Free PMC article. Review.
-
Mammalian iron sulfur cluster biogenesis: From assembly to delivery to recipient proteins with a focus on novel targets of the chaperone and co-chaperone proteins.IUBMB Life. 2022 Jul;74(7):684-704. doi: 10.1002/iub.2593. Epub 2022 Jan 25. IUBMB Life. 2022. PMID: 35080107 Free PMC article. No abstract available.
-
The assembly of succinate dehydrogenase: a key enzyme in bioenergetics.Cell Mol Life Sci. 2019 Oct;76(20):4023-4042. doi: 10.1007/s00018-019-03200-7. Epub 2019 Jun 24. Cell Mol Life Sci. 2019. PMID: 31236625 Free PMC article. Review.
References
-
- Harding A.E. Early onset cerebellar ataxia with retained tendon reflexes: a clinical and genetic study of a disorder distinct from Friedreich's ataxia. J. Neurol. Neurosurg. Psychiatry. 1981;44:503–508. doi:10.1136/jnnp.44.6.503. - DOI - PMC - PubMed
-
- Campuzano V., Montermini L., Molto M.D., Pianese L., Cossee M., Cavalcanti F., Monros E., Rodius F., Duclos F., Monticelli A., et al. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi:10.1126/science.271.5254.1423. - DOI - PubMed
-
- Becker E., Richardson D.R. Frataxin: its role in iron metabolism and the pathogenesis of Friedreich's ataxia. Int. J. Biochem. Cell Biol. 2001;33:1–10. doi:10.1016/S1357-2725(00)00067-4. - DOI - PubMed
-
- Adamec J., Rusnak F., Owen W.G., Naylor S., Benson L.M., Gacy A.M., Isaya G. Iron-dependent self-assembly of recombinant yeast frataxin: implications for Friedreich's ataxia. Am. J. Hum. Genet. 2000;67:549–562. doi:10.1086/303056. - DOI - PMC - PubMed
-
- Puccio H., Simon D., Cossee M., Criqui-Filipe P., Tiziano F., Melki J., Hindelang C., Matyas R., Rustin P., Koenig M. Mouse models for Friedreich's ataxia exhibit cardiomyopathy, sensory nerve defect and Fe–S enzyme deficiency followed by intramitochondrial iron deposits. Nat. Genet. 2001;27:181–186. doi:10.1038/84818. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

