Protein unfolding and degradation by the AAA+ Lon protease
- PMID: 22162032
- PMCID: PMC3324771
- DOI: 10.1002/pro.2013
Protein unfolding and degradation by the AAA+ Lon protease
Abstract
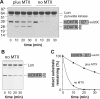
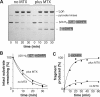
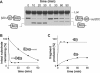
AAA+ proteases employ a hexameric ring that harnesses the energy of ATP binding and hydrolysis to unfold native substrates and translocate the unfolded polypeptide into an interior compartment for degradation. What determines the ability of different AAA+ enzymes to unfold and thus degrade different native protein substrates is currently uncertain. Here, we explore the ability of the E. coli Lon protease to unfold and degrade model protein substrates beginning at N-terminal, C-terminal, or internal degrons. Lon has historically been viewed as a weak unfoldase, but we demonstrate robust and processive unfolding/degradation of some substrates with very stable protein domains, including mDHFR and titin(I27) . For some native substrates, Lon is a more active unfoldase than related AAA+ proteases, including ClpXP and ClpAP. For other substrates, this relationship is reversed. Thus, unfolding activity does not appear to be an intrinsic enzymatic property. Instead, it depends on the specific protease and substrate, suggesting that evolution has diversified rather than optimized the protein unfolding activities of different AAA+ proteases.
Copyright © 2011 The Protein Society.
Figures
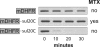


Similar articles
-
Single-molecule denaturation and degradation of proteins by the AAA+ ClpXP protease.Proc Natl Acad Sci U S A. 2009 Nov 17;106(46):19340-5. doi: 10.1073/pnas.0910484106. Epub 2009 Nov 5. Proc Natl Acad Sci U S A. 2009. PMID: 19892734 Free PMC article.
-
Engineering fluorescent protein substrates for the AAA+ Lon protease.Protein Eng Des Sel. 2013 Apr;26(4):299-305. doi: 10.1093/protein/gzs105. Epub 2013 Jan 28. Protein Eng Des Sel. 2013. PMID: 23359718 Free PMC article.
-
Degrons in protein substrates program the speed and operating efficiency of the AAA+ Lon proteolytic machine.Proc Natl Acad Sci U S A. 2009 Nov 3;106(44):18503-8. doi: 10.1073/pnas.0910392106. Epub 2009 Oct 19. Proc Natl Acad Sci U S A. 2009. PMID: 19841274 Free PMC article.
-
ClpXP, an ATP-powered unfolding and protein-degradation machine.Biochim Biophys Acta. 2012 Jan;1823(1):15-28. doi: 10.1016/j.bbamcr.2011.06.007. Epub 2011 Jun 27. Biochim Biophys Acta. 2012. PMID: 21736903 Free PMC article. Review.
-
Slicing a protease: structural features of the ATP-dependent Lon proteases gleaned from investigations of isolated domains.Protein Sci. 2006 Aug;15(8):1815-28. doi: 10.1110/ps.052069306. Protein Sci. 2006. PMID: 16877706 Free PMC article. Review.
Cited by
-
Structures of the human LONP1 protease reveal regulatory steps involved in protease activation.Nat Commun. 2021 May 28;12(1):3239. doi: 10.1038/s41467-021-23495-0. Nat Commun. 2021. PMID: 34050165 Free PMC article.
-
Biomolecular mechanisms for signal differentiation.iScience. 2021 Nov 17;24(12):103462. doi: 10.1016/j.isci.2021.103462. eCollection 2021 Dec 17. iScience. 2021. PMID: 34927021 Free PMC article.
-
Slippery substrates impair ATP-dependent protease function by slowing unfolding.J Biol Chem. 2013 Nov 29;288(48):34729-35. doi: 10.1074/jbc.M113.512533. Epub 2013 Oct 22. J Biol Chem. 2013. PMID: 24151080 Free PMC article.
-
Identification of a Degradation Signal Sequence within Substrates of the Mitochondrial i-AAA Protease.J Mol Biol. 2017 Mar 24;429(6):873-885. doi: 10.1016/j.jmb.2017.02.009. Epub 2017 Feb 16. J Mol Biol. 2017. PMID: 28214511 Free PMC article.
-
N-terminomics identifies widespread endoproteolysis and novel methionine excision in a genome-reduced bacterial pathogen.Sci Rep. 2017 Sep 11;7(1):11063. doi: 10.1038/s41598-017-11296-9. Sci Rep. 2017. PMID: 28894154 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

