Genotype-phenotype correlations in Lesch-Nyhan disease: moving beyond the gene
- PMID: 22157001
- PMCID: PMC3270957
- DOI: 10.1074/jbc.M111.317701
Genotype-phenotype correlations in Lesch-Nyhan disease: moving beyond the gene
Abstract
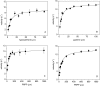
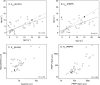
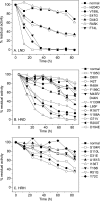
Lesch-Nyhan disease and its attenuated variants are caused by mutations in the HPRT1 gene, which encodes the purine recycling enzyme hypoxanthine-guanine phosphoribosyltransferase. The mutations are heterogeneous, with more than 400 different mutations already documented. Prior efforts to correlate variations in the clinical phenotype with different mutations have suggested that milder phenotypes typically are associated with mutants that permit some residual enzyme function, whereas the most severe phenotype is associated with null mutants. However, multiple exceptions to this concept have been reported. In the current studies 44 HPRT1 mutations associated with a wide spectrum of clinical phenotypes were reconstructed by site-directed mutagenesis, the mutant enzymes were expressed in vitro and purified, and their kinetic properties were examined toward their substrates hypoxanthine, guanine, and phosphoribosylpyrophosphate. The results provide strong evidence for a correlation between disease severity and residual catalytic activity of the enzyme (k(cat)) toward each of its substrates as well as several mechanisms that result in exceptions to this correlation. There was no correlation between disease severity and the affinity of the enzyme for its substrates (K(m)). These studies provide a valuable model for understanding general principles of genotype-phenotype correlations in human disease, as the mechanisms involved are applicable to many other disorders.
Figures

Similar articles
-
New biomarkers for early diagnosis of Lesch-Nyhan disease revealed by metabolic analysis on a large cohort of patients.Orphanet J Rare Dis. 2015 Jan 23;10:7. doi: 10.1186/s13023-014-0219-0. Orphanet J Rare Dis. 2015. PMID: 25612837 Free PMC article.
-
Genotype-phenotype correlations in neurogenetics: Lesch-Nyhan disease as a model disorder.Brain. 2014 May;137(Pt 5):1282-303. doi: 10.1093/brain/awt202. Epub 2013 Aug 22. Brain. 2014. PMID: 23975452 Free PMC article. Review.
-
Clinical severity in Lesch-Nyhan disease: the role of residual enzyme and compensatory pathways.Mol Genet Metab. 2015 Jan;114(1):55-61. doi: 10.1016/j.ymgme.2014.11.001. Epub 2014 Nov 8. Mol Genet Metab. 2015. PMID: 25481104 Free PMC article.
-
Genotypic and phenotypic spectrum in attenuated variants of Lesch-Nyhan disease.Mol Genet Metab. 2014 Aug;112(4):280-5. doi: 10.1016/j.ymgme.2014.05.012. Epub 2014 May 28. Mol Genet Metab. 2014. PMID: 24930028 Free PMC article. Review.
-
Lesch-Nyhan disease.Nucleosides Nucleotides Nucleic Acids. 2008 Jun;27(6):559-63. doi: 10.1080/15257770802135745. Nucleosides Nucleotides Nucleic Acids. 2008. PMID: 18600504
Cited by
-
Small Duplication of HPRT 1 Gene May Be Causative For Lesh-Nyhan Disease in Iranian Patients.Iran J Child Neurol. 2015 Winter;9(1):103-6. Iran J Child Neurol. 2015. PMID: 25767547 Free PMC article.
-
Mutations in POMGNT1 cause non-syndromic retinitis pigmentosa.Hum Mol Genet. 2016 Apr 15;25(8):1479-88. doi: 10.1093/hmg/ddw022. Epub 2016 Jan 28. Hum Mol Genet. 2016. PMID: 26908613 Free PMC article.
-
Clinical utility gene card for: Lesch-Nyhan syndrome--update 2013.Eur J Hum Genet. 2013 Oct;21(10). doi: 10.1038/ejhg.2012.304. Epub 2013 Jan 16. Eur J Hum Genet. 2013. PMID: 23321622 Free PMC article. No abstract available.
-
New biomarkers for early diagnosis of Lesch-Nyhan disease revealed by metabolic analysis on a large cohort of patients.Orphanet J Rare Dis. 2015 Jan 23;10:7. doi: 10.1186/s13023-014-0219-0. Orphanet J Rare Dis. 2015. PMID: 25612837 Free PMC article.
-
Genotype-phenotype correlations in neurogenetics: Lesch-Nyhan disease as a model disorder.Brain. 2014 May;137(Pt 5):1282-303. doi: 10.1093/brain/awt202. Epub 2013 Aug 22. Brain. 2014. PMID: 23975452 Free PMC article. Review.
References
-
- Jinnah H. A., De Gregorio L., Harris J. C., Nyhan W. L., O'Neill J. P. (2000) The spectrum of inherited mutations causing HPRT deficiency. 75 new cases and a review of 196 previously reported cases. Mutat. Res. 463, 309–326 - PubMed
-
- Jinnah H. A., Harris J. C., Nyhan W. L., O'Neill J. P. (2004) The spectrum of mutations causing HPRT deficiency. An update. Nucleosides Nucleotides Nucleic Acids 23, 1153–1160 - PubMed
-
- Jolly D. J., Okayama H., Berg P., Esty A. C., Filpula D., Bohlen P., Johnson G. G., Shively J. E., Hunkapillar T., Friedmann T. (1983) Isolation and characterization of a full-length expressible cDNA for human hypoxanthine phosphoribosyltransferase. Proc. Natl. Acad. Sci. U.S.A. 80, 477–481 - PMC - PubMed
-
- Keebaugh A. C., Sullivan R. T., Thomas J. W. (2007) NISC Comparative Sequencing Program, Gene duplication and inactivation in the HPRT gene family. Genomics 89, 134–142 - PubMed
-
- 5 Jinnah H. A., Visser J. E., Harris J. C., Verdu A., Larovere L., Ceballos-Picot I., Gonzalez-Alegre P., Neychev V., Torres R. J., Dulac O., Desguerre I., Schretlen D. J., Robey K. L., Barabas G., Bloem B. R., Nyhan W., De Kremer R., Eddey G.E., Puig J. G., Reich S. G., and Lesch-Nyhan Disease International Study Group (2006) Delineation of the motor disorder of Lesch-Nyhan disease. Brain 129, 1201–1217 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

