Regulation of polysome assembly on the endoplasmic reticulum by a coiled-coil protein, p180
- PMID: 22156060
- PMCID: PMC3326322
- DOI: 10.1093/nar/gkr1197
Regulation of polysome assembly on the endoplasmic reticulum by a coiled-coil protein, p180
Abstract
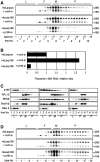
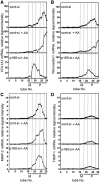
A coiled-coil microtubule-bundling protein, p180, was originally identified as one of the ribosome receptor candidates on the rough endoplasmic reticulum (ER) and is highly expressed in secretory tissues. Recently, we reported that p180 plays crucial roles in upregulating collagen biosynthesis, mainly by facilitating ribosome association on the ER. Here, we provide evidence that p180 is required to form translationally active polysome/translocon complexes on the ER. Assembly of highly-developed polysomes on the ER was severely perturbed upon loss of p180. p180 associates with polysome/translocon complexes through multiple contact sites: it was coimmunoprecipitated with the translocon complex independently of ribosomes, while it can also bind to ribosomal large subunit specifically. The responsible domain of p180 for membrane polysome assembly was identified in the C-terminal coiled-coil region. The degree of ribosome occupation of collagen and fibronectin mRNAs was regulated in response to increased traffic demands. This effect appears to be exerted in a manner specific for a specified set of mRNAs. Collectively, our data suggest that p180 is required to form translationally active polysome/translocon complexes on the ER membrane, and plays a pivotal role in highly efficient biosynthesis on the ER membrane through facilitating polysome formation in professional secretory cells.
Figures





Similar articles
-
Enhancement of procollagen biosynthesis by p180 through augmented ribosome association on the endoplasmic reticulum in response to stimulated secretion.J Biol Chem. 2010 Sep 24;285(39):29941-50. doi: 10.1074/jbc.M109.094607. Epub 2010 Jul 20. J Biol Chem. 2010. PMID: 20647306 Free PMC article.
-
Expansion of the trans-Golgi network following activated collagen secretion is supported by a coiled-coil microtubule-bundling protein, p180, on the ER.Exp Cell Res. 2010 Feb 1;316(3):329-40. doi: 10.1016/j.yexcr.2009.11.009. Epub 2009 Nov 27. Exp Cell Res. 2010. PMID: 19932094
-
Component of splicing factor SF3b plays a key role in translational control of polyribosomes on the endoplasmic reticulum.Proc Natl Acad Sci U S A. 2019 May 7;116(19):9340-9349. doi: 10.1073/pnas.1901742116. Epub 2019 Apr 19. Proc Natl Acad Sci U S A. 2019. PMID: 31004060 Free PMC article.
-
Compartmentation of the rough endoplasmic reticulum.Mol Cell Biochem. 1986 Jun;71(1):3-18. doi: 10.1007/BF00219323. Mol Cell Biochem. 1986. PMID: 2425244 Review.
-
In vitro and tissue culture methods for analysis of translation initiation on the endoplasmic reticulum.Methods Enzymol. 2007;431:47-60. doi: 10.1016/S0076-6879(07)31004-5. Methods Enzymol. 2007. PMID: 17923230 Review.
Cited by
-
ER ribosomal-binding protein 1 regulates blood pressure and potassium homeostasis by modulating intracellular renin trafficking.J Biomed Sci. 2023 Feb 19;30(1):13. doi: 10.1186/s12929-023-00905-7. J Biomed Sci. 2023. PMID: 36803854 Free PMC article.
-
Role of LARP6 and nonmuscle myosin in partitioning of collagen mRNAs to the ER membrane.PLoS One. 2014 Oct 1;9(10):e108870. doi: 10.1371/journal.pone.0108870. eCollection 2014. PLoS One. 2014. PMID: 25271881 Free PMC article.
-
An RNA-centric dissection of host complexes controlling flavivirus infection.Nat Microbiol. 2019 Dec;4(12):2369-2382. doi: 10.1038/s41564-019-0518-2. Epub 2019 Aug 5. Nat Microbiol. 2019. PMID: 31384002 Free PMC article.
-
Transmembrane and coiled-coil domain family 1 is a novel protein of the endoplasmic reticulum.PLoS One. 2014 Jan 14;9(1):e85206. doi: 10.1371/journal.pone.0085206. eCollection 2014. PLoS One. 2014. PMID: 24454821 Free PMC article.
-
Axonal endoplasmic reticulum tubules control local translation via P180/RRBP1-mediated ribosome interactions.Dev Cell. 2024 Aug 19;59(16):2053-2068.e9. doi: 10.1016/j.devcel.2024.05.005. Epub 2024 May 29. Dev Cell. 2024. PMID: 38815583 Free PMC article.
References
-
- Diehn M, Eisen MB, Botstein D, Brown PO. Large-scale identification of secreted and membrane-associated gene products using DNA microarrays. Nat. Genet. 2000;25:58–62. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

